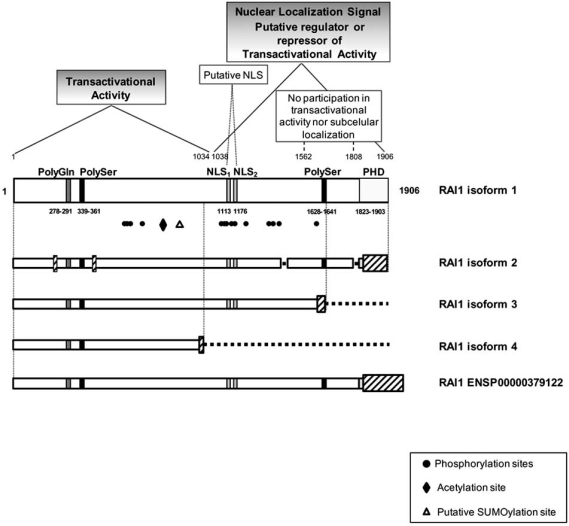
Fig. (1).
Structure of RAI1. By in silico analyses, several domains have been found for RAI1: a polyglutamine tract at the N-terminal of the protein, two polyserine domains, a PHD domain at the C-terminal of RAI1 and two putative nuclear localization signals (NLS). RAI1 protein structure is schematically represented, and the domains described are showed with their localization in the amino acidic sequence. By the evaluation of the wild type and mutant proteins associated to SMS, two main domains were found: the N-terminal half of RAI1 is responsible for the transactivational activity and C-terminal half beginning in residue 1038 is responsible for nuclear localization [34]. The RAI1 PTMs found are depicted with black figures while the putative sites are represented by white figures. The phosphorylation sites (1Thr and 13Ser) found in HeLa cells are represented with black circles. The reported acetylation site is depicted with a black diamond. The putative SUMOylation site with the highest score is represented with a white triangle.
Below, the other four isoforms described for human RAI1 are represented. These isoforms differ from the canonical one by the change of several residues (depicted in slanted rectangles) and also by some fragments missed (depicted by discontinued lines). Isoform 2 is the most similar to the canonical, while in isoform 3 are missing the second polyserine tract and PHD domain. Isoform 4 is the shortest containing only the first half of RAI1 protein, not encompassing NLSs and being comparable to the truncated proteins found in some SMS patients. The longest variant of RAI1 (ENSP00000379122) is a protein reported by the online database of eukaryotic genomes, Ensembl, which has a length of 1993 amino acids and has complete homology with the isoform 1 until amino acid 1855.