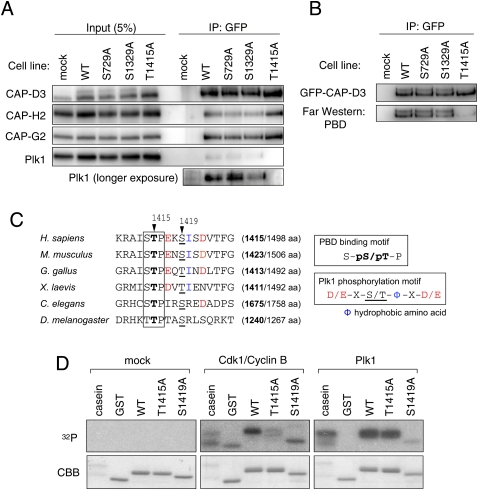
Figure 2.
Identification of phosphorylation sites on CAP-D3. (A) Immunoprecipitation of Plk1 with CAP-D3 is abolished in a Thr 1415 nonphosphorylatable mutant. Mitotic cell extracts prepared from cells stably expressing the indicated version of GFP-tagged CAP-D3 were subjected to immunoprecipitation analysis with antibodies to GFP and were immunoblotted with the antibodies indicated. (B) The binding of PBD to CAP-D3 is lost in the T1415A mutant. The indicated series of GFP-tagged CAP-D3 proteins were immunoprecipitated (top panel) and analyzed by Far-Western analysis with PBD (bottom panel). Note that, like the endogenous protein, GFP-CAP-D3 can be detected as two major bands in mitotic extracts from the wild-type and S729A and S1329A mutant cells, but not the T1415A cells. (C) A PBD-binding motif in CAP-D3. Among three candidate sites, Thr 1415 and Ser 1419 fall within a PBD-binding motif (boxed) and the Plk1 consensus phosphorylation site (color-coded), respectively. Equivalent regions from orthologous proteins from different species are aligned, highlighting the evolutionary conservation of these motifs. The numbers in brackets indicate positions of the Thr 1415-equivalent threonine (bold) in the full amino acid length of CAP-D3 protein. (D) Cdk1 and Plk1 mediate phosphorylation of Thr 1415 and Ser 1419, respectively, in vitro. A series of polypeptides corresponding to a partial fragment of CAP-D3 that encompasses the prospective phosphorylation sites were incubated with mock (control), Cdk1/Cyclin B, or Plk1. (Top panels) Incorporation of 32P was detected by autoradiography. (Bottom panels) Coomassie Brilliant Blue staining (CBB) verifies that equivalent amounts of substrate appear in each lane.