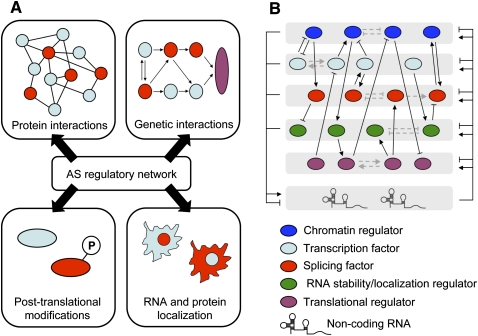
FIGURE 2.
Regulatory outcomes in alternative splicing networks and cross-talk between factors acting at multiple layers of gene regulation. (A) Coordinated alternative splicing events from an alternative splicing network (isoforms regulated by a specific splicing factor are displayed in red in the top and bottom panels) could result in the modification of protein–protein interactions (top left panel), modulation of genetic interactions within biological pathways (top right panel), altered post-translational modification sites in targeted proteins (bottom left panel), and altered RNA or protein localization in the cell (bottom right panel). Both single candidate gene studies and high-throughput approaches can be used to understand the functional consequences of these regulatory events. (B) Cross-regulation (black lines and arrows) has been observed between various classes of trans-acting regulators, such as chromatin-modifying proteins (blue), splicing factors (red), transcription factors (light blue), RNA stability or localization factors (green), translational regulators (purple), and non-coding RNAs. Cross-regulatory interactions have also been observed between members acting in the same process (gray dashed lines and arrows). The diagram displays both known regulatory events as well as theoretical events. A detailed understanding of how each of these factors interact and regulate each other will be essential for a complete understanding of the underlying regulatory networks they control.