Abstract
Pseudogenes have long been labeled as “junk” DNA, failed copies of genes that arise during the evolution of genomes. However, recent results are challenging this moniker; indeed, some pseudogenes appear to harbor the potential to regulate their protein-coding cousins. Far from being silent relics, many pseudogenes are transcribed into RNA, some exhibiting a tissue-specific pattern of activation. Pseudogene transcripts can be processed into short interfering RNAs that regulate coding genes through the RNAi pathway. In another remarkable discovery, it has been shown that pseudogenes are capable of regulating tumor suppressors and oncogenes by acting as microRNA decoys. The finding that pseudogenes are often deregulated during cancer progression warrants further investigation into the true extent of pseudogene function. In this review, we describe the ways in which pseudogenes exert their effect on coding genes and explore the role of pseudogenes in the increasingly complex web of noncoding RNA that contributes to normal cellular regulation.
Keywords: pseudogenes, functional, noncoding RNA, transcription, RNA
INTRODUCTION
The human genome, like that of other mammals, is littered with a variety of repetitive elements and noncoding genes. One such element is the pseudogene, a poor facsimile of an original protein-coding gene that has lost the ability to produce a functional protein (Mighell et al. 2000). Because they do not code for proteins, pseudogenes are often assumed to be nonfunctional and labeled as “junk DNA.” While some pseudogenes are transcriptionally silent, others are active, raising the question of whether their noncoding transcripts are a spurious use of cellular energy or instead harnessed by the cell to regulate coding genes (Balakirev and Ayala 2003). This question is particularly pertinent given the recent flurry of evidence suggesting that long noncoding RNAs play a critical role in regulating genomic function (Mattick and Makunin 2006; Guttman et al. 2009; Caley et al. 2010).
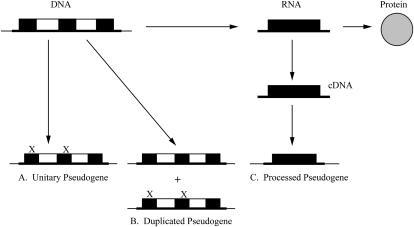
Pseudogenes can arise through a variety of mechanisms. Spontaneous mutations in a coding gene that prevent either transcription or translation of the gene (Fig. 1A) result in the formation of “unitary” pseudogene (Zhang et al. 2010). Duplicated pseudogenes are created via tandem duplication or uneven crossing-over (Mighell et al. 2000). These duplicated genes lose their protein-coding potential due to the loss of promoters or enhancers or crippling mutations such as frameshifts or premature stop codons (Fig. 1B). However, they do tend to retain their characteristic intron-exon structure. In contrast, retrotransposed or “processed” pseudogenes (PPs) are produced when an mRNA transcript is reverse-transcribed and integrated into the genome at a new location (Fig. 1C), and therefore, they do not normally contain introns (Maestre et al. 1995; D'Errico et al. 2004). Other common features of PPs are their polyA tracts and direct repeats at either end of the pseudogene (Maestre et al. 1995; D'Errico et al. 2004). The retrotransposition of mRNAs into the genome appears to be mediated by long interspersed nuclear element 1 (L1) (Esnault et al. 2000; Ding et al. 2006), and the transcriptional activity of the resulting PP depends on whether the integration event occurs close to another promoter (Zheng et al. 2007). The collection of processed pseudogenes in the human genome has been generated from just 10% of the coding genes (Ohshima et al. 2003; Zhang et al. 2003). Highly expressed housekeeping genes are more likely to produce PPs, as are other highly transcribed shorter RNAs (Gonçalves et al. 2000). This is exemplified by the small number of genes encoding ribosomal proteins that account for ∼20% of human PPs (Zhang et al. 2002).
FIGURE 1.
Types of pseudogene. (A) Mutation of existing genes gives rise to unitary pseudogenes. (B) Duplicated pseudogenes are produced following mutation of copied genes. (C) Reverse transcription of mRNA into cDNA followed by retrotransposition into genomic DNA leads to the generation of processed pseudogenes. Filled boxes represent exons; open boxes represent introns; X represents a crippling mutation that ablates protein-coding potential.
The term pseudogene was coined in 1977, when Jacq and co-workers discovered a version of the gene coding for 5S rRNA that was truncated but retained homology with the active gene in Xenopus laevis (Jacq et al. 1977). During the following two decades, pseudogenes were discovered in a sporadic fashion (Mighell et al. 2000). The acceleration in next-generation sequencing technologies coupled with the monumental human genome project has yielded the full genome sequences of a range of organisms, permitting much more thorough analyses of pseudogene prevalence (Ohshima et al. 2003; Torrents et al. 2003; Zhang et al. 2003). Remarkably, pseudogenes are almost as numerous as coding genes, with predictions ranging from 10,000 to 20,000 human pseudogenes (Zhang and Gerstein 2004). The majority of human pseudogenes are PPs (Ohshima et al. 2003; Torrents et al. 2003; Zhang et al. 2003), while the number of unitary pseudogenes in the human genome is <100 (Zhang et al. 2010). Pseudogenes are present in a wide range of species, including plants (Loguercio and Wilkins 1998; Benovoy and Drouin 2006), bacteria (Ochman and Davalos 2006) [though they are not as numerous in unicellular organisms (Lawrence et al. 2001)], insects (Ramos-Onsins and Aguadé 1998; Harrison et al. 2003), and nematode worms (Harrison et al. 2001), but they are particularly numerous in mammals (Zhang and Gerstein 2004).
EVOLUTION AND CONSERVATION OF PSEUDOGENES
Pseudogenes are sometimes considered to represent “neutral sequence,” in which mutations that accumulate are neither selected for or against (Li et al. 1981). However, this premise relies on the assumption that pseudogenes are functionally inert. There is recent evidence that some pseudogenes are functionally active, and therefore, studying their evolution and conservation could support a functional role and give insight into their potential mechanism of action.
The synonymous to nonsynonymous substitution rate (KA/KS) is a measure of the proportion of mutations in DNA sequence that also alter amino acid sequence; it is often used to assess whether a sequence is under evolutionary constraint (Hurst 2002; Torrents et al. 2003). It has been reported recently (for review, see Balakirev and Ayala 2003) that synonymous point mutations occur far more frequently than do nonsynonymous base changes in the Drosophila Est-6 pseudogene (Balakirev et al. 2003). In chickens, it has been observed that, within the multiple IglV and IghV pseudogenes, the number of stop codons contained in the “coding” sequence is far lower than would be expected were nucleotide substitutions occurring at random; in addition, the majority of stop codons introduced by point mutations are then “corrected” and eliminated by further point mutations within the same codon (Rothenfluh et al. 1995). This feature, which is also observed in VH pseudogenes in mice (Schiff et al. 1985), may indicate either that some presumed pseudogenes are, in fact, protein-coding, or that the conservation of open reading frames plays a role in any putative function of pseudogenes involved in somatic gene rearrangements (Balakirev and Ayala 2003).
A recent report described a region of the 3′ UTR of the PTENP1 pseudogene that shared over 95% sequence homology with the PTEN coding gene (Poliseno et al. 2010). The functional implications of this pattern and high degree of conservation are discussed in further detail below.
Pseudogenes gradually accumulate mutations, and the number of mutations can give us an estimate of their age. Fascinatingly, the appearance of Alu elements in Old World primates coincided with the peak of processed pseudogene generation and subsequent radiation of primates ∼40 million years ago (Ohshima et al. 2003; Zhang et al. 2003). Conservation of pseudogenes across different species has also been observed. Analysis of the rhesus macaque major histocompatibility complex (MHC) extended class II region revealed two pseudogenes that were found to be homologous to the human HIV TAT-specific factor-1-like and zinc finger-like pseudogenes, which was suggestive of evolutionary conservation (Sudbrak et al. 2003). Investigations by Podlaha and colleagues (Podlaha and Zhang 2004) demonstrated that the Makorin1-p1 pseudogene is conserved across Mus musculus and Mus pahari strains. This prompted a genome-wide survey for pseudogenes conserved between humans and mice in which human pseudogenes, along with their parent genes, were compared with the corresponding mouse orthologues and their pseudogenes (Svensson et al. 2006). Interestingly, many of the pseudogenes examined were found to have very few mutations within the regulatory regions they shared with their parent genes, which might suggest that these regulatory regions are of importance to the pseudogene and that the pseudogene may be functional. Of the groups of genes and pseudogenes examined, sequence analysis suggested that 30 of them represented pseudogenes that were present in both mice and humans and had arisen before the two species diverged. Comparison of transcribed human pseudogenes shows that ∼50% are conserved with rhesus monkey, but only 3% are conserved in mouse (Khachane and Harrison 2009). Analysis of these pseudogenes showed that, in spite of a KA/KS substitution rate indicative of noncoding RNAs, the levels of GC and the rate of mutation in these pseudogenes is constrained relative to the intergenic regions that surround them (Khachane and Harrison 2009). The collective evidence that some pseudogenes exhibit sequence conservation hints at a potential functional role in the organisms that harbor them.
TRANSCRIPTION OF PSEUDOGENES
Most pseudogenes lose the ability to be transcribed, either due to mutations in their promoter, or (in the case of PPs) integration into silent regions of the genome. Making accurate measurements of pseudogene transcription is complicated by the similarity they share with their parent genes (Ruud et al. 1999; Harper et al. 2003). However, there are numerous examples of pseudogenes that are transcribed, including pseudogenes for the tumor suppressor PTEN (whose transcripts are more numerous than the parent gene) (Fujii et al. 1999), the adrenal steroid hydroxylase P450c21A (Bristow et al. 1993), human leukocyte interferon (Goeddel et al. 1981), GAPDH (Tso et al. 1985), glucocerebrosidase (Sorge et al. 1990), and Oct4 (Redshaw and Strain 2010). Microarray technology permits analysis of pseudogene transcription on a much larger scale. Estimates of the proportion of transcribed human pseudogenes vary from 2% to 20% (Yano et al. 2004; Harrison et al. 2005; Zheng et al. 2005, 2007).
Investigating the pattern of transcription across tissues and cell lines can give insight into potential functionality. Other noncoding RNAs exhibit tissue specific expression patterns and have also been shown to have functional roles, including antisense RNAs (Dahary et al. 2005; Katayama et al. 2005), intergenic transcripts and long noncoding RNAs (Bertone et al. 2004; Cheng et al. 2005; Guttman et al. 2009) and miRNAs (Zhang 2008). In a survey of transcription in the ENCODE regions of the genome, 14 pseudogenes were found to be transcribed (Zheng et al. 2007). Five of these were transcribed exclusively in the testes, and a further four were also active in testes and other tissues (Zheng et al. 2007). This pattern of transcription is consistent with previous results, suggesting a possible biological significance for testes-specific pseudogene transcription (Kleene et al. 1998; Reymond et al. 2002). There are also several examples of pseudogenes whose spatio-temporal expression pattern is different from that of their parent gene (Olsen and Schechter 1999; Elliman et al. 2006).
Specific changes in pseudogene expression can also occur under different physiological conditions, including diseases such as diabetes (Chiefari et al. 2010) and cancer (Suo et al. 2005; Zou et al. 2009; Poliseno et al. 2010). Examples of dynamic pseudogene transcription have been observed in other organisms. Robust pseudogene transcription has been measured in Mycobacterium leprae, the organism that causes leprosy, with levels of specific transcripts varying during the process of infection (Suzuki et al. 2006). Induction of stress in Arabidopsis thaliana leads to changes in expression of numerous genes and pseudogenes (Zeller et al. 2009).
The transcriptional activity of a pseudogene will, in part, depend on the promoter it is utilizing. Some have their own promoters, while others use the promoters of nearby genes (Harrison et al. 2005; Vinckenbosch et al. 2006). A processed pseudogene is largely at the mercy of its integration site when it comes to promoter activity. Therefore, the difference in transcription pattern between the pseudogene and its parent gene does not necessarily reflect a functional role but may be instead merely the result of being driven by a new promoter. If the latter were true, then one could predict that the act of transcribing a pseudogene would be evolutionarily neutral (or even selected against to conserve cellular energy). Analysis of conserved transcribed pseudogenes shows that ∼50% are indeed conserved across millions of years of primate evolution (though far fewer are conserved between species more distant to human, such as rodents) (Khachane and Harrison 2009). That transcription of some pseudogenes is tissue-specific, dynamic, and has been maintained over millennia suggests that their transcripts may play some functional role in cells. There is no substitute, however, for functional experiments to test whether pseudogenes and their transcripts have a direct active role.
EVIDENCE FOR PSEUDOGENE FUNCTION
Many multicellular organisms appear to conserve the presence of pseudogenes and impose constraint on the sequences themselves. Various unicellular organisms, on the other hand, actively expel genes that have become pseudogenized (Kuo and Ochman 2010). The question remains, then, of what is the potential benefit to mammals and other complex organisms in retaining and arguably embracing genes that have lost protein-coding potential? One proposed function of pseudogenes is to act as a source of genetic diversity, for example, in the generation of antibodies or antigen variation (Balakirev and Ayala 2003). However, it is in the context of noncoding RNA that pseudogenes offer potentially much more dynamic mechanisms to regulate ongoing nuclear processes. In the last decade, a new layer of complexity has been revealed in the regulation of gene expression and nuclear function. Many noncoding RNAs, both long and short, have been shown to regulate a variety of processes in cells. The noncoding RNAs produced from some pseudogenes also appear to use a variety of fascinating mechanisms to control gene function.
There is evidence that pseudogenes and their parent genes form regulatory pairs that can influence each other. Specific knockdown of the ATP-binding cassette (ABC) transporter pseudogene ABCC6P1 causes a decrease in the expression of ABCC6 mRNA (Piehler et al. 2008). Oct4, a pluripotency-associated transcription factor, has several known pseudogenes. Overexpression of a putative pseudogene transcript (Oct4P1) leads to inhibition of mesenchymal stem cell differentiation while stimulating proliferation (Lin et al. 2007). An inverse correlation was observed between BRAF pseudogene transcription and the quantity of BRAF mutations during the progression of papillary thyroid carcinoma (Zou et al. 2009). Forced transcription of the BRAF pseudogene stimulated MAP kinase signaling, transformed NIH3T3 cells in culture, and induced tumors in nude mice (Zou et al. 2009). Fascinatingly, the Xist noncoding RNA, which mediates dosage compensation by coating the inactive X chromosome in mammals and mediating epigenetic repression, may have evolved by pseudogenization of a protein-coding ancestor called Lnx3 (Duret et al. 2006).
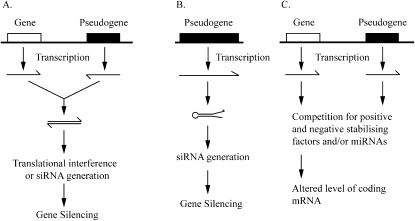
It has long been hypothesized that antisense pseudogene transcripts could combine with sense genic transcripts to regulate levels of expression (Fig. 2A) (McCarrey and Riggs 1986). Examples of this mechanism have now been reported. Knockdown of an RNA antisense to an Oct4 pseudogene leads to an apparent increase in levels of both Oct4 and two of its pseudogenes (Hawkins and Morris 2010). Simultaneous transcription of both neural nitric oxide synthase (nNOS) and a related pseudogene in the same neurons of Lymnaea stagnalis (a species of snail) leads to the formation of a duplex between the two strands and a reduction in nNOS translation (Korneev et al. 1999).
FIGURE 2.
Potential mechanisms of pseudogene function. (A) Antisense RNA generated from pseudogenes can combine with sense-stranded mRNA from a homologous coding gene and either inhibit translation or lead to the formation of siRNAs that can inhibit expression of the coding gene. (B) Similarly, siRNAs can be generated from hairpin structures within folded pseudogene transcripts. (C) RNA from coding genes and their homologous pseudogenes can compete for trans-acting stability factors and degradation by miRNAs. When the levels of pseudogene transcripts are altered, this, in turn, affects the stability of the coding gene mRNA and therefore, expression levels.
In two landmark reports, it was shown that in mouse oocytes, portions of many pseudogene transcripts are processed into small interfering RNAs (siRNAs) (Tam et al. 2008; Watanabe et al. 2008). These siRNAs were derived either from pseudogenes with internal secondary structure (Fig. 2B), or from dsRNAs composed of sense and antisense transcripts (pseudogene-pseudogene and pseudogene-coding mRNA combinations were observed). The loss of Dicer (a protein necessary for the production of siRNAs) led to a decrease in the levels of pseudogene-derived siRNAs and an increase in the level of coding gene mRNAs with homology with the siRNA sequences (Tam et al. 2008; Watanabe et al. 2008). This suggests that the siRNAs generated from the dsRNA are capable of repressing gene expression. For example, siRNAs generated from a hairpin structure in the Au76 pseudogene RNA were able to inhibit expression from the parent coding gene Rangap1 (Watanabe et al. 2008). Several siRNAs were detected bearing sequence similarity to the histone deacetylase complex, HDAC1 (Tam et al. 2008). All of the siRNA sequences were derived from a series of HDAC1 pseudogenes (none from the HDAC1 gene itself), yet upon Dicer knockout, the levels of HDAC1 mRNA increased, suggesting that the coding gene is regulated by a RNA-induced silencing complex (RISC)-based process. In another case, the siRNAs were generated from regions of dsRNA formed between Ppp4r1 mRNA and an antisense RNA generated from a pseudogene with 90% homology (Watanabe et al. 2008). The siRNAs produced from this pairing appear to repress the Ppp4r1 gene. A similar survey in a species of rice shows that a small proportion of pseudogenes are transcribed and processed into siRNAs after pairing with the coding gene or a paralogous pseudogene transcript (Guo et al. 2009). These findings suggest a potential mechanism by which antisense transcripts may operate. However, it remains to be seen whether similar processes occur in mammalian somatic cells.
Several groups have proposed that pseudogenes may interfere with factors that regulate mRNA stability (Hirotsune et al. 2003; Piehler et al. 2008; Chiefari et al. 2010). It is known that the stability of a given mRNA depends on cis-acting sequences and their interaction with trans-acting molecules (Ross 1996). If a pseudogene and parent coding gene harbor similar cis-acting sequences, then it is feasible that they may compete for the same trans-acting elements (Fig. 2C). Thus, regulated changes in pseudogene transcription could lead to altered coding-gene mRNA stability and, therefore, expression. Such a role was suggested for the imprinted Makorin1-p1 pseudogene in modulating the expression of its imprinted parent gene Makorin-1 (Hirotsune et al. 2003). Reduction of Makorin-p1 RNA led to a drop in mRNA from the protein-coding gene and a significant disease phenotype in mice (Hirotsune et al. 2003). This finding remains controversial, however, as a subsequent study showed that the pseudogene is, in fact, transcriptionally silent, and neither gene is imprinted (Gray et al. 2006). Decreased production of high mobility group A1 (HMGA1) protein can lead to the deregulation of the insulin receptor (INSR) gene and subsequent development of type 2 diabetes mellitus (Brunetti et al. 2001). In two recently tested type 2 diabetes patients, the low level of HMGA1 was also associated with a high level of HMGA1 pseudogene (HMGA1-p) mRNA (Chiefari et al. 2010). Knockdown of HMGA1-p RNA led to partially restored HMGA1 mRNA levels, suggesting that the two transcripts may compete for a trans-acting stability factor.
miRNAs are a class of noncoding RNA that affect mRNA stability. Their specificity and function is mediated by base pairing with the target (primarily at the 3′ UTR); their primary effect is to cause degradation of the mRNA and therefore reduce levels of expression (Bartel 2009; Guo et al. 2010). In a recent report, a gene-pseudogene pair was shown to be co-regulated by the same miRNAs (Poliseno et al. 2010). PTEN is a tumor suppressor that is often mutated at one allele (while wild type at the second allele) at cancer presentation (Salmena et al. 2008). The severity and susceptibility to cancer is influenced by the dosage of PTEN (Alimonti et al. 2010). Therefore, maintaining precise levels of PTEN protein is critical for preventing oncogenesis. PTENP1, a pseudogene of the PTEN tumor suppressor, is transcribed at a high level (Fujii et al. 1999). As described above, the 5′-most region of the PTENP1 3′ UTR is ∼95% similar in sequence to the 3′ UTR of PTEN (Poliseno et al. 2010). A number of miRNAs are capable of targeting this region on both the gene and pseudogene. Knockdown of the PTENP1 transcript leads to reduced levels of PTEN mRNA and protein and inhibition of growth; conversely, transfecting cells with the 3′ UTR of PTENP1 caused PTEN expression to increase (Poliseno et al. 2010). This suggests that the PTENP1 pseudogene acts as a “miRNA decoy,” binding to and thereby reducing the effective cellular concentration of miRNAs, therefore allowing PTEN to escape miRNA-mediated repression. A functional link between the PTEN/PTENP1 pair is consistent with the findings that their levels are often correlated in prostate cancer samples and that focal deletions containing PTENP1 occur frequently in sporadic colon cancer cases (Poliseno et al. 2010). A similar relationship was also demonstrated between the oncogene KRAS and its pseudogene KRASP1 (Poliseno et al. 2010). Interestingly, the ability of the HMGA1-p pseudogene to destabilize HMGA1 mRNA depends on the 3′ UTR region (Chiefari et al. 2010), suggesting that both positive and negative stabilization factors are able to interact competitively with the untranslated regions of genes and pseudogenes to regulate expression output.
CONCLUSIONS
Caution must be exercised when interpreting the results of functional experiments on pseudogenes. In some cases, what appears to be a nontranslated pseudogene can, in fact, code for truncated proteins (Kandouz et al. 2004; Zhang et al. 2006). Nevertheless, the evidence that some pseudogenes can exert regulatory effects on their protein-coding cousins is mounting. Such functions appear to be mediated by noncoding RNAs produced from active pseudogenes. While not all pseudogenes (or even all transcribed pseudogenes) will have biological functions, it is likely that, where an unexpected regulatory benefit results from the formation of a pseudogene, the effect will be conserved. For the large part, pseudogenes have been overlooked in the quest to understand the biology of health and disease, to the extent that pseudogene probes are often absent from commercially available microarrays. As evidence emerges that pseudogenes are deregulated in disease, and indeed that their deregulation can contribute to diseases such as diabetes and cancer, the prevalent attitude that they are nonfunctional relics is slowly changing. With the advent of affordable next- generation sequencing, the study of transcriptomics, and in particular, pseudogenes (and other transcribed noncoding elements), should experience a quantum leap forward. In the coming decade, the extent and mechanisms of pseudogene function should become clearer.
ACKNOWLEDGMENTS
R.C.P. is supported by a grant from Action Medical Research; D.P.C. is supported by a grant from the Dunhill Medical Trust. We also thank the Cancer and Polio Research Fund and Oxford Brookes University for financial support.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2658311.
REFERENCES
- Alimonti A, Carracedo A, Clohessy JG, Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ, Brogi E, et al. 2010. Subtle variations in Pten dose determine cancer susceptibility. Nat Genet 42: 454–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakirev ES, Ayala FJ 2003. Pseudogenes: Are they “junk” or functional DNA? Annu Rev Genet 37: 123–151 [DOI] [PubMed] [Google Scholar]
- Balakirev ES, Chechetkin VR, Lobzin VV, Ayala FJ 2003. DNA polymorphism in the beta-Esterase gene cluster of Drosophila melanogaster. Genetics 164: 533–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benovoy D, Drouin G 2006. Processed pseudogenes, processed genes, and spontaneous mutations in the Arabidopsis genome. J Mol Evol 62: 511–522 [DOI] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce T, Rozowsky J, Urban A, Zhu X, Rinn J, Tongprasit W, Samanta M, Weissman S, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Bristow J, Gitelman SE, Tee MK, Staels B, Miller WL 1993. Abundant adrenal-specific transcription of the human P450c21A “pseudogene”. J Biol Chem 268: 12919–12924 [PubMed] [Google Scholar]
- Brunetti A, Manfioletti G, Chiefari E, Goldfine ID, Foti D 2001. Transcriptional regulation of human insulin receptor gene by the high-mobility group protein HMGI(Y). FASEB J 15: 492–500 [DOI] [PubMed] [Google Scholar]
- Caley D, Pink R, Trujillano D, Carter D 2010. Long noncoding RNAs, chromatin, and development. ScientificWorldJournal 10: 90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Chiefari E, Iiritano S, Paonessa F, Le Pera I, Arcidiacono B, Filocamo M, Foti D, Liebhaber SA, Brunetti A 2010. Pseudogene-mediated posttranscriptional silencing of HMGA1 can result in insulin resistance and type 2 diabetes. Nat Commun 1: 1–7 [DOI] [PubMed] [Google Scholar]
- Dahary D, Elroy-Stein O, Sorek R 2005. Naturally occurring antisense: transcriptional leakage or real overlap? Genome Res 15: 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Errico I, Gadaleta G, Saccone C 2004. Pseudogenes in metazoa: Origin and features. Brief Funct Genomics Proteomics 3: 157–167 [DOI] [PubMed] [Google Scholar]
- Ding W, Lin L, Chen B, Dai J 2006. L1 elements, processed pseudogenes and retrogenes in mammalian genomes. IUBMB Life 58: 677–685 [DOI] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312: 1653–1655 [DOI] [PubMed] [Google Scholar]
- Elliman SJ, Wu I, Kemp DM 2006. Adult tissue-specific expression of a Dppa3-derived retrogene represents a postnatal transcript of pluripotent cell origin. J Biol Chem 281: 16–19 [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T 2000. Human LINE retrotransposons generate processed pseudogenes. Nat Genet 24: 363–367 [DOI] [PubMed] [Google Scholar]
- Fujii GH, Morimoto AM, Berson AE, Bolen JB 1999. Transcriptional analysis of the PTEN/MMAC1 pseudogene, psiPTEN. Oncogene 18: 1765–1769 [DOI] [PubMed] [Google Scholar]
- Goeddel DV, Leung DW, Dull TJ, Gross M, Lawn RM, McCandliss R, Seeburg PH, Ullrich A, Yelverton E, Gray PW 1981. The structure of eight distinct cloned human leukocyte interferon cDNAs. Nature 290: 20–26 [DOI] [PubMed] [Google Scholar]
- Gonçalves I, Duret L, Mouchiroud D 2000. Nature and structure of human genes that generate retropseudogenes. Genome Res 10: 672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Wilson A, Fortin PJ, Nicholls RD 2006. The putatively functional Mkrn1-p1 pseudogene is neither expressed nor imprinted, nor does it regulate its source gene in trans. Proc Natl Acad Sci 103: 12039–12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhang Z, Gerstein MB, Zheng D 2009. Small RNAs originated from pseudogenes: cis- or trans-acting? PLoS Comput Biol 5: e1000449 doi: 10.1371/journal.pcbi.1000449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin M, Feldser D, Huarte M, Zuk O, Carey B, Cassady J, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper LV, Hilton AC, Jones AF 2003. RT-PCR for the pseudogene-free amplification of the glyceraldehyde-3-phosphate dehydrogenase gene (gapd). Mol Cell Probes 17: 261–265 [DOI] [PubMed] [Google Scholar]
- Harrison PM, Echols N, Gerstein MB 2001. Digging for dead genes: An analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome. Nucleic Acids Res 29: 818–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Milburn D, Zhang Z, Bertone P, Gerstein M 2003. Identification of pseudogenes in the Drosophila melanogaster genome. Nucleic Acids Res 31: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Zheng D, Zhang Z, Carriero N, Gerstein M 2005. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res 33: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins PG, Morris KV 2010. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcr 1: 165–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A 2003. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423: 91–96 [DOI] [PubMed] [Google Scholar]
- Hurst LD 2002. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet 18: 486 doi: 10.1016/S0168-9525(02)02722-1 [DOI] [PubMed] [Google Scholar]
- Jacq C, Miller J, Brownlee G 1977. A pseudogene structure in 5S DNA of Xenopus laevis. Cell 12: 109–120 [DOI] [PubMed] [Google Scholar]
- Kandouz M, Bier A, Carystinos GD, Alaoui-Jamali MA, Batist G 2004. Connexin43 pseudogene is expressed in tumor cells and inhibits growth. Oncogene 23: 4763–4770 [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Khachane AN, Harrison PM 2009. Assessing the genomic evidence for conserved transcribed pseudogenes under selection. BMC Genomics 10: 435 doi: 10.1186/1471-2164-10-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene KC, Mulligan E, Steiger D, Donohue K, Mastrangelo MA 1998. The mouse gene encoding the testis-specific isoform of Poly(A) binding protein (Pabp2) is an expressed retroposon: Intimations that gene expression in spermatogenic cells facilitates the creation of new genes. J Mol Evol 47: 275–281 [DOI] [PubMed] [Google Scholar]
- Korneev SA, Park JH, O'Shea M 1999. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J Neurosci 19: 7711–7720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CH, Ochman H 2010. The extinction dynamics of bacterial pseudogenes. PLoS Genet 6: e1001050 doi: 10.1371/journal.pgen.1001050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Hendrix RW, Casjens S 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol 9: 535–540 [DOI] [PubMed] [Google Scholar]
- Li WH, Gojobori T, Nei M 1981. Pseudogenes as a paradigm of neutral evolution. Nature 292: 237–239 [DOI] [PubMed] [Google Scholar]
- Lin H, Shabbir A, Molnar M, Lee T 2007. Stem cell regulatory function mediated by expression of a novel mouse Oct4 pseudogene. Biochem Biophys Res Commun 355: 111–116 [DOI] [PubMed] [Google Scholar]
- Loguercio LL, Wilkins TA 1998. Structural analysis of a hmg-coA-reductase pseudogene: Insights into evolutionary processes affecting the hmgr gene family in allotetraploid cotton (Gossypium hirsutum L.). Curr Genet 34: 241–249 [DOI] [PubMed] [Google Scholar]
- Maestre J, Tchénio T, Dhellin O, Heidmann T 1995. mRNA retroposition in human cells: Processed pseudogene formation. EMBO J 14: 6333–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J, Makunin I 2006. Non-coding RNA. Hum Mol Genet 15: R17–R29 [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Riggs AD 1986. Determinator-inhibitor pairs as a mechanism for threshold setting in development: A possible function for pseudogenes. Proc Natl Acad Sci 83: 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mighell AJ, Smith NR, Robinson PA, Markham AF 2000. Vertebrate pseudogenes. FEBS Lett 468: 109–114 [DOI] [PubMed] [Google Scholar]
- Ochman H, Davalos LM 2006. The nature and dynamics of bacterial genomes. Science 311: 1730–1733 [DOI] [PubMed] [Google Scholar]
- Ohshima K, Hattori M, Yada T, Gojobori T, Sakaki Y, Okada N 2003. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol 4: R74 doi: 10.1186/gb-2003-4-11-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen MA, Schechter LE 1999. Cloning, mRNA localization and evolutionary conservation of a human 5-HT7 receptor pseudogene. Gene 227: 63–69 [DOI] [PubMed] [Google Scholar]
- Piehler AP, Hellum M, Wenzel JJ, Kaminski E, Haug KB, Kierulf P, Kaminski WE 2008. The human ABC transporter pseudogene family: Evidence for transcription and gene-pseudogene interference. BMC Genomics 9: 165 doi: 10.1186/1471-2164-9-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlaha O, Zhang J 2004. Nonneutral evolution of the transcribed pseudogene Makorin1-p1 in mice. Mol Biol Evol 21: 2202–2209 [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins S, Aguadé M 1998. Molecular evolution of the Cecropin multigene family in Drosophila. Functional genes vs. pseudogenes. Genetics 150: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redshaw Z, Strain AJ 2010. Human haematopoietic stem cells express Oct4 pseudogenes and lack the ability to initiate Oct4 promoter-driven gene expression. J Negat Results Biomed 9: 2 doi: 10.1186/1477-5751-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Marigo V, Yaylaoglu MB, Leoni A, Ucla C, Scamuffa N, Caccioppoli C, Dermitzakis ET, Lyle R, Banfi S, et al. 2002. Human chromosome 21 gene expression atlas in the mouse. Nature 420: 582–586 [DOI] [PubMed] [Google Scholar]
- Ross J 1996. Control of messenger RNA stability in higher eukaryotes. Trends Genet 12: 171–175 [DOI] [PubMed] [Google Scholar]
- Rothenfluh HS, Blanden RV, Steele EJ 1995. Evolution of V genes: DNA sequence structure of functional germline genes and pseudogenes. Immunogenetics 42: 159–171 [DOI] [PubMed] [Google Scholar]
- Ruud P, Fodstad O, Hovig E 1999. Identification of a novel cytokeratin 19 pseudogene that may interfere with reverse transcriptase-polymerase chain reaction assays used to detect micrometastatic tumor cells. Int J Cancer 80: 119–125 [DOI] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP 2008. Tenets of PTEN tumor suppression. Cell 133: 403–414 [DOI] [PubMed] [Google Scholar]
- Schiff C, Milili M, Fougereau M 1985. Functional and pseudogenes are similarly organized and may equally contribute to the extensive antibody diversity of the IgVHII family. EMBO J 4: 1225–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge J, Gross E, West C, Beutler E 1990. High level transcription of the glucocerebrosidase pseudogene in normal subjects and patients with Gaucher disease. J Clin Invest 86: 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbrak R, Reinhardt R, Hennig S, Lehrach H, Günther E, Walter L 2003. Comparative and evolutionary analysis of the rhesus macaque extended MHC class II region. Immunogenetics 54: 699–704 [DOI] [PubMed] [Google Scholar]
- Suo G, Han J, Wang X, Zhang J, Zhao Y, Dai J 2005. Oct4 pseudogenes are transcribed in cancers. Biochem Biophys Res Commun 337: 1047–1051 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Nakata N, Bang PD, Ishii N, Makino M 2006. High-level expression of pseudogenes in Mycobacterium leprae. FEMS Microbiol Lett 259: 208–214 [DOI] [PubMed] [Google Scholar]
- Svensson Ö, Arvestad L, Lagergren J 2006. Genome-wide survey for biologically functional pseudogenes. PLoS Comput Biol 2: e46 doi: 10.1371/journal.pcbi.0020046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents D, Suyama M, Zdobnov E, Bork P 2003. A genome-wide survey of human pseudogenes. Genome Res 13: 2559–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso JY, Sun XH, Kao TH, Reece KS, Wu R 1985. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: Genomic complexity and molecular evolution of the gene. Nucleic Acids Res 13: 2485–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci 103: 3220–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Yano Y, Saito R, Yoshida N, Yoshiki A, Wynshaw-Boris A, Tomita M, Hirotsune S 2004. A new role for expressed pseudogenes as ncRNA: Regulation of mRNA stability of its homologous coding gene. J Mol Med 82: 414–422 [DOI] [PubMed] [Google Scholar]
- Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, Weigel D, Laubinger S 2009. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 58: 1068–1082 [DOI] [PubMed] [Google Scholar]
- Zhang C 2008. MicroRNomics: A newly emerging approach for disease biology. Physiol Genomics 33: 139–147 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Gerstein M 2004. Large-scale analysis of pseudogenes in the human genome. Curr Opin Genet Dev 14: 328–335 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Harrison P, Gerstein M 2002. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res 12: 1466–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Harrison PM, Liu Y, Gerstein M 2003. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res 13: 2541–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang X, Li M, Han J, Chen B, Wang B, Dai J 2006. NANOGP8 is a retrogene expressed in cancers. FEBS J 273: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Zhang ZD, Frankish A, Hunt T, Harrow J, Gerstein M 2010. Identification and analysis of unitary pseudogenes: historic and contemporary gene losses in humans and other primates. Genome Biol 11: R26 doi: 10.1186/gb-2010-11-3-r26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Zhang Z, Harrison PM, Karro J, Carriero N, Gerstein M 2005. Integrated pseudogene annotation for human chromosome 22: Evidence for transcription. J Mol Biol 349: 27–45 [DOI] [PubMed] [Google Scholar]
- Zheng D, Frankish A, Baertsch R, Kapranov P, Reymond A, Choo SW, Lu Y, Denoeud F, Antonarakis SE, Snyder M, et al. 2007. Pseudogenes in the ENCODE regions: Consensus annotation, analysis of transcription, and evolution. Genome Res 17: 839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Baitei EY, Alzahrani AS, Al-Mohanna F, Farid NR, Meyer B, Shi Y 2009. Oncogenic activation of MAP kinase by BRAF pseudogene in thyroid tumors. Neoplasia 11: 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]