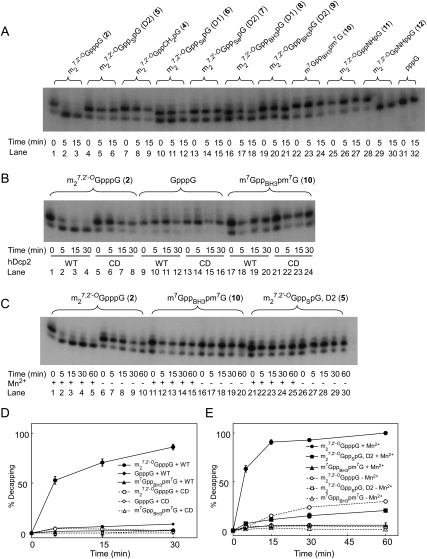
FIGURE 2.
In vitro hydrolysis of capped oligonucleotides by hDcp2 analyzed on RNA sequencing gels. Oligonucleotides were synthesized from Nco1-cut pluc-A60 template with T7 polymerase in the presence of [α-32P]GTP and various cap dinucleotides. After treatment with hDcp2 for the indicated times, samples were loaded on a 10% RNA sequencing gel as described in Materials and Methods. (A) Ten RNA transcripts synthesized in the presence of either no cap dinucleotide (lanes 31,32) or the indicated cap dinucleotide were treated with 0.6 μg of hDcp2 for the indicated times and analyzed by electrophoresis and autoradiography. Samples in lanes labeled 0 min did not receive hDcp2. The quantification of decapping at 5 min and 15 min is given in Table 1. (B) Oligonucleotides synthesized in the presence of the indicated cap dinucleotides were subjected to digestion with 0.4 μg of either wild-type (WT) or catalytic dead (CD) hDcp2 for the indicated times and subjected to electrophoresis as in A. (C) Oligonucleotides synthesized in the presence of the indicated cap dinucleotides were subjected to digestion with 1 μg of WT hDcp2 for the indicated times, with or without 0.5 mM Mn2+ in the reaction mixture as indicated, and analyzed by electrophoresis and autoradiography. (D) Quantification of gel in B. (E) Quantification of gel in C. Decapping was calculated as the % loss in the upper band, normalized by the radioactivity in the upper plus lower bands. The data represent the means and SEM of three experiments. Error bars smaller than the symbols are not shown.