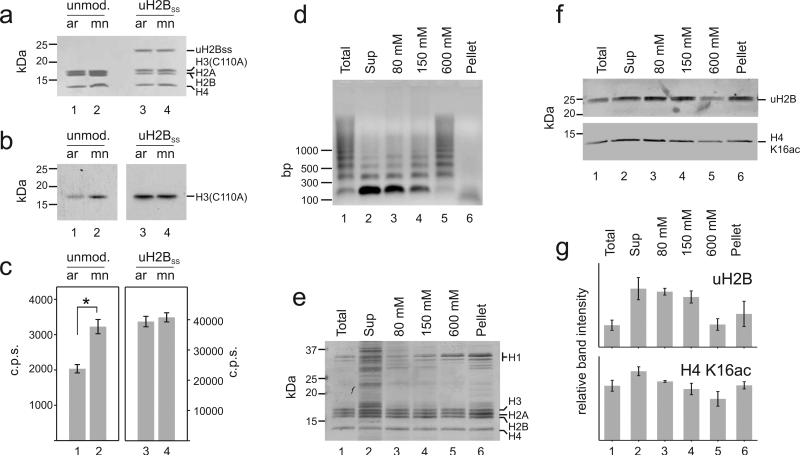
Figure 2. H2B ubiquitylation increases chromatin accessibility in vitro and in vivo.
a-c, Unmodified arrays (ar) (lane 1) and mono-nucleosomes (mn) (lane 2), or H2B ubiquitylated arrays (lane 3) and mono-nucleosomes (lane 4) were used as substrates for hDot1L methyltransferase assays with 3H-SAM at 1 mM Mg2+. Histones were separated by SDS-PAGE, (a) stained with Coomassie Brilliant Blue and (b) probed for 3H-methyl incorporation by fluorography. c, Quantification of methylation was performed by p81 filter binding followed by liquid scintillation counting. Student's two tailed t-test: *: p = 0.0001; Error bars, SEM (n=9). The fluorography represents two different exposures of the film: 24 h, left panel and 3 h, right panel. For verification of the modification site and full gels, see Supplementary Fig. 3. d-g Chromatin from micrococcal nuclease digested nuclei of NIH/3T3 fibroblasts was successively extracted with increasing concentrations of NaCl. d, DNA purified from MNase digested nuclei (lane 1), from the supernatant after centrifugation of the nuclei (lane 2), from 80, 150 and 600 mM salt extracted chromatin (lanes 3, 4, 5) and from the insoluble pellet (lane 6) was separated on an agarose gel and stained with Ethidium bromide. Histones were acid extracted and equal amounts were separated by SDS-PAGE and either (e) stained with Coomassie Brilliant Blue or (f) analyzed by Western blotting with antibodies against uH2B and H4 K16ac. For full gels see Supplementary Fig. 3. g, uH2B and H4 K16ac levels from two independent experiments were quantified by densitometry. Error bars, SEM (n=2).