Abstract
We have developed a complete system for the isotopic labeling, fractionation, and automated quantification of differentially expressed peptides that significantly facilitates candidate biomarker discovery. We describe a new stable mass tagging reagent pair, 12C6- and 13C6-phenylisocyanate (PIC), that offers significant advantages over currently available tags. Peptides are labeled predominantly at their amino termini and exhibit elution profiles that are independent of label isotope. Importantly, PIC-labeled peptides have unique neutral-mass losses upon CID fragmentation that enable charge state and label isotope identification and, thereby, decouple the sequence identification from the quantification of candidate biomarkers. To exploit these properties, we have coupled peptide fractionation protocols with a Thermo LTQ-XL LC-MS2 data acquisition strategy and a suite of automated spectrum analysis software that identifies quantitative differences between labeled samples. This approach, dubbed the PICquant platform, is independent of protein sequence identification and excludes unlabeled peptides that otherwise confound biomarker discovery. Application of the PICquant platform to a set of complex clinical samples showed that the system allows rapid identification of peptides that are differentially expressed between control and patient groups.
Keywords: PICquant, mass spectrometry, quantification, phenylisocyanate, stable mass tag, urine
Introduction
Identification and quantification of differentially expressed and/or post-translationally modified (PTM) proteins in disease specimens represent the fundamental goals of proteomic biomarker discovery. Over the past two decades, proteomic approaches using mass spectrometry (MS) have rapidly evolved such that thousands of peptides from hundreds of proteins can be identified from a single liquid chromatography/tandem mass spectrometry (LC-MS/MS) analysis. Recent efforts have also explored a growing number of strategies that enable quantitative analysis of proteins and peptides in complex mixtures. These strategies can be categorized as either “mass tag” chemical labeling or “label-free” approaches.
Though stable isotope mass tagging represents a promising approach to quantification of peptides and proteins,1–4 a variety of limitations hinder current mass tagging strategies. For example, mass tags such as the ICAT reagent label only a subset of peptides in a mixture and require a separate purification step.5–7 Esterification with deuterated methanol is complicated by the variable number of deuterium atoms attached per peptide (i.e. 3, 6, or 9 m/z, or more) and the observation that deuterated peptides display an increased hydrophobicity.8 Mass tagging approaches with 18O water9,10 are limited by the small mass difference between the isotope labels and exchange rates that can be dependent upon local structure and environment. An isobaric mass tag, iTraq, has been developed that indiscriminately labels amino groups with one of eight stable mass tags.11 However, quantification is achieved through analysis of small fragmentation products, currently possible only with select mass spectral instruments, such as the Applied Biosystems QSTAR XL Pro, and is complicated by reagent impurities.4 Indiscriminate amine labeling also means that many peptides contain two or more labels, potentially complicating sequence identification. Isobaric mass tagging has also been developed using two different isotope tags to derivatize the peptide termini in a complementary fashion such that the total added mass is equivalent.12 This approach shares the advantage of other isobaric approaches in not doubling the complexity of the labeled sample, but it involves multiple labeling steps, labels only peptides ending with lysine, and generates fragmentation ion doublets that can inhibit peptide sequencing. Finally, the SILAC methodology13–16 that involves metabolically labeling cell cultures with dual isotopic versions of an essential amino acid, such as 13C6-Arg or 13C6-Lys, has proven to be a powerful technique. However, as this approach is limited to systems where the proteins are synthesized in growth media containing label, it is not generally applicable to tissues, body fluids or clinical applications.
These various mass-tagging strategies typically require sequence identification of peptide ions before quantification. This restricts analysis to the 20 to 30% of the acquired MS2 scans that can be confidently matched to a peptide sequence via database search algorithms.17–19 While some of the unmatched MS2 scans represent non-peptide ions derived from chemical or electronic sources, many other scans derive from peptide ions but remain unidentified because of poor signal quality or poor fragmentation. Still other peptide ions remain unmatched despite high quality MS2 scans because the peptide contains amino acid polymorphisms, post-translational modifications, or splice variants that are not anticipated by the search algorithm, or the parent protein is not included within the sequence database. Regardless, if the quantitative analysis of the sample is restricted to only those ions that are confidently matched to a peptide sequence, a large portion of the peptide ions will elude discovery.
An alternative to mass-tagging approaches, label-free quantification strategies have taken two general approaches.4,20,21 The first, usually requiring a high-resolution mass spectrometer, extracts the chromatographic elution profiles of all the ions observed in the MS1 scans. Differences in these ion profiles across multiple runs of different samples are used to quantify changes in the relative peptide abundances. Because this strategy does not require MS2 acquisitions, it effectively increases the dynamic range of the peptide detection since it does not have the problem of under-sampling peptide ions observed in MS1 scans. Unlike most mass tagging methods, this approach uncouples peptide quantification from the sequence identification processes that may instead occur later in a targeted manner. The success of this approach is strongly dependent on the consistency of the peptide chromatographic elution over multiple acquisitions and typically employs a number of sophisticated alignment methods that attempt to correct for all of the various chromatographic fluctuations.22–25
A second label-free approach, commonly referred to as spectral counting, assumes that the frequency at which a peptide is identified is proportional to its abundance in the sample.26 A number of studies have demonstrated that spectral counting approaches have a wider dynamic range with respect to measurements based on signal intensity and provide reproducible protein quantification when spectral counts and signal-to-noise levels are high.27,28 However, these efforts have two intrinsic disadvantages. First, despite algorithms that attempt to take into account MS2 under-sampling and database search errors,29 spectral counting efforts ultimately rely upon peptide identifications that are completely dependent upon peptide sequence identification, relying on pre-existing knowledge of protein sequences and post-translational modifications that may be lacking from the search library. Second, while low abundance peptides are likely to be of the greatest experimental interest, these peptides are likely to be rarely sampled for MS2 analysis; yet spectral counting cannot quantify with statistical significance peptide identifications with fewer than 4 acquisitions.20,28,30,31
The work presented here describes our effort to circumvent many of these disadvantages of the different quantitative proteomic approaches by coupling a unique data acquisition strategy using 12C6- and 13C6-phenylisocyanate (PIC) mass tags with a suite of open-source scripts. Collectively termed “PICquant”, this platform takes advantage of the unique neutral mass losses generated upon CID fragmentation of PIC-labeled peptides to quantify peptide ions automatically and anonymously, without the requirement for sequence identification. Focused sequence identification strategies are performed only on the relatively few differentially-expressed peptides. Application of the PICquant workflow to a clinical project, urine samples from patients scheduled for a biopsy of a suspicious breast lump, demonstrated efficient identification and quantification of differentially expressed peptides across a multiply fractionated sample.
Materials and Methods
Details of the urine sample preparation, immunoblots, mass spectrometry and data analysis are provided in Supplemental Material.
Phenylisocyanate Isotopomer Labels
13C6-phenylisocyanate (PIC-H) at 99+% isotopic purity (Cat # 603597) was obtained from Isotech of Sigma-Aldrich (St. Louis, MO) and was stored either in anhydrous conditions at room temperature or as a 100 mM acetonitrile solution at −20°C. Conventional 12C6-phenylisocyanate (PIC-L) was obtained from Acros Organics (Morris Plains, NJ). For the PIC-labeling reactions, 100 mM triethylammonium acetate TEAA buffer was used for the protein sample because it does not have a free amine that can react with the phenylisocyanate. Acetic acid was used to bring the pH of the TEAA buffer to 7.5 in order to preferentially label the α-amine of peptides. The phenylisocyanate label from its 100 mM stock solution was added to the tryptic peptides at a 10:1 molar ratio and the reaction was quenched after 10 minutes at room temperature by the addition of ammonium bicarbonate.
Urine Sample Preparation
Urine was collected with appropriate consent and IRB approval from patients with a suspicious breast mass. Patient files were reviewed retrospectively to identify controls (five subjects with benign breast disease) or patients (five subjects with invasive adenocarcinoma). Protein from 15 to 30 mL of urine from each control and patient (10 total samples) was denatured and reduced with dithiothreitol, carboxyamidomethylated with iodoacetamide, and then passed through a 50 kDa cutoff Amicon Ultra centrifugal filter (Millipore, Billerica, MA) to separate the high-molecular weight proteins and a 3 kDa cutoff to desalt and concentrate. Retentates were washed 3× with 100 mM TEAA buffer at pH 7.5, protein concentrations were measured by Bradford assay (Bio-Rad, Hercules, CA) and normalized pooled control and patient samples were then prepared for both the low-molecular (3–50 kDa) and high-molecular (>50 kDa) fractions.
For isoelectric focusing (IEF) fractionation, 60 µg of trypsinized protein from the low-molecular fraction of the control and patient pools were incubated with PIC-L or PIC-H, respectively. Both pools were combined, prepared for IEF according to manufacturer recommendation, and fractionated on Immobiline IPG Drystrip pH 3–10. The IEF gel strip was cut into 13 (1 cm) pieces, the peptides extracted and subjected to a ZipTip (Millipore, Billerica, MA) clean-up.
For SDS-PAGE fractionation, a 40 µg aliquot from both the high-molecular and low-molecular protein patient and control pools were fractionated on 12% SDS-PAGE gels that were then cut into slices. Slightly modifying a procedure previously described,32 the gel slices were chopped into 1 mm cubes, incubated with trypsin overnight, extracted into a TEAA pH 7.5 buffer, and then incubated with either PIC-L for the patient sample or PIC-H for the control sample. After quenching the reactions with ammonium bicarbonate, the control and patient samples were combined for analysis.
Mass Spectrometry
The Thermo LTQ-XL ion trap mass spectrometer (Thermo, San Jose, CA) was operated in the data dependent mode with an Agilent 1100 HPLC system split to nano-flow. The acquisition duty cycle consisted of an initial MS1centroid scan with a mass range of 300–2000 m/z for all experiments, except for repeat experiments of SDS-PAGE gel samples for which the mass range was set at 500–1000 m/z. The 5 most abundant ions were sequentially selected for a Zoom MS1 scan acquired in profile with a width of 20 m/z centered on the precursor ion. Each Zoom MS1 scan was followed by a MS2 CID spectrum of that same precursor. After repeating for each of the top five precursor ions, the cycle repeated. The duty cycle for this data acquisition cycle of 11 mass spectral scans was about 3 s.
Data Processing and Analysis
Data sets were handled using a Perl script, dubbed MAZIE, written in our lab that accurately determines peptide charge and monoisotopic mass for each MS2 scan precursor ion by analyzing the preceding Zoom MS1 scan33 and then generates a concatenated DTA file used for searching with the OMSSA engine.34 MAZIE is distributed under the Creative Commons License, and is available, together with its dependencies, at http://faculty.virginia.edu/templeton. The MS2 data was searched as both a tryptic and semi-tryptic digest against a composite database containing the human refseq database, (ftp.ncbi.nih.gov/refseq), and the reversed protein sequences generated by an in-house Perl script. Search parameters were optimized as described previously,33 with the mass of both the precursor and fragment ions treated as monoisotopic with an m/z tolerance of 0.3 Da and 0.5 Da, respectively. Tryptic and semi-tryptic search results for each data file were merged by retaining the matched OMSSA hit, if any, for each MS2 scan that had the most confident (lowest) E-value and then loaded into a MySQL database.
MySQL 5.1 (http://www.mysql.com) was installed on a Macintosh Pro computer as part of the MAMP package (http://www.mamp.info) that includes Apache 2 and PHP5.
The Perl script “PICquant” was used to identify and quantify ion pairs differentially labeled with PIC-L and PIC-H, as described in Results and Discussion. It is included as a module of the MAZIE algorithm and is available as described above.
Results and Discussion
The primary objective of the PIC labeling strategy and PICquant analysis platform is to uncover potential biomarker candidates by identifying differentially expressed proteins/peptides in complex biological samples. To develop and validate this labeling protocol, we used urine from patients scheduled for a biopsy of a suspicious breast lump, obtained at UVA under an Institutional Review Board approved protocol. While the clinical samples represent typical clinical samples that might be analyzed, we make no claims to the clinical diagnostic utility of any proteins or other information derived from this study. Samples from case (invasive ductal adenocarcinoma) and control (no malignant disease) patients were pooled into two separate groups, with urine from six patients approximately equally represented on a protein mass basis in each pool.
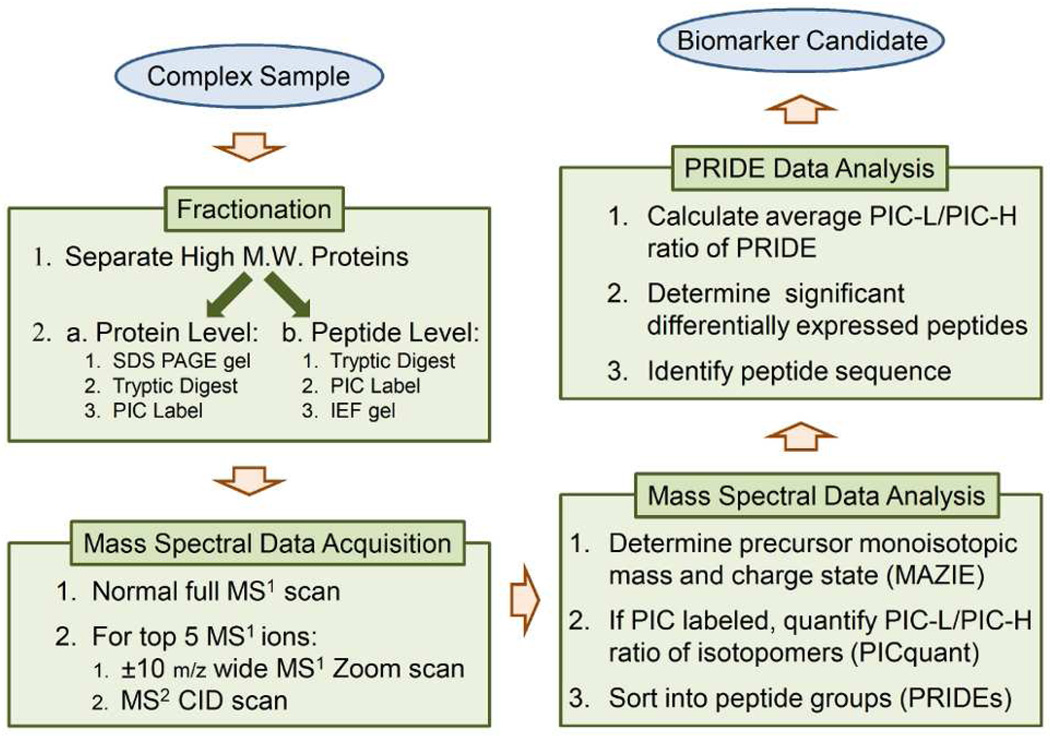
The flowchart in Figure 1 outlines the general approach of PICquant. Because fractionation greatly facilitates a deep analysis, we used two complementary techniques to fractionate samples at either the protein (SDS-PAGE gel) or the peptide (IEF) level after a size filtration had already segregated albumin and other large proteins. The high-abundance albumin otherwise obscures peptides that are present at much lower concentrations. As detailed in Materials and Methods, we prepared differentially-labeled pooled samples from patients and controls and then analyzed paired fractions from both the SDS-PAGE and IEF gel techniques.
Figure 1.
An overview of the PICquant platform workflow.
Phenylisocyanate (PIC) Labeling
Phenylisocyanate (PIC) labeling of peptides exploits the difference in nucleophilicity between the N-terminal α-amine versus the ε-amine on the lysine side chain.35 At neutral pH, the N-terminal α-amine of peptides reacts with isocyanates 100 times faster than does the ε-amino group of lysine,36 thereby enabling relatively specific labeling of each peptide at only one location. Other investigators have used a D5-labeled form of phenylisocyanate as an isotope tag for model peptide and protein systems.37 However, because deuterium-labeled peptides elute earlier than their non-labeled counterpart during reverse-phase chromatography,38 quantification of peptide isotopomers is significantly more difficult. To overcome this limitation, we employed a 13C6-PIC label. The 6 Da isotope mass difference is sufficient to resolve peptide isotopomers with charge states up to +4 on a low-resolution LTQ.
The efficiency and specificity of the PIC labeling was quantified using a bovine serum albumin (BSA) tryptic digest (see Supplemental Data). Mass spectral data was acquired both with and without natural 12C-PIC (PIC-L) labeling. Because the ionization efficiency of the PIC-labeled derivatives was most likely altered by modification of the N-terminal free amine, the completeness of PIC labeling for a given peptide was determined by the reduction in the observed ion peaks across its different charge states. The average across the 17 most prominent tryptic peptides suggests an N-terminal PIC-labeling efficiency of roughly 85% (see Supplemental Table 1). The specificity of the reaction was demonstrated by the presence of the single-labeled PIC derivative of all of these peptides at the N-terminal α-amine. Though more complete labeling could be forced by increasing the reaction time and/or the PIC concentration, this also increases the population of peptides doubled-labeled at the ε-amino group of a lysine (not shown). Supplemental Material contains further details about the PIC labeling reaction.
Data Acquisition
The PICquant platform requires accurate monoisotopic mass and charge state information for the precursor peptide ions. Typically acquired in centroid mode, the nominal MS1 scan of a Thermo LTQ mass spectrometer does not have the mass resolution necessary to accurately quantify the intensities associated with specific isotopic peaks of a peptide ion. However, we recently described a software algorithm, MAZIE,33 that accurately determines the monoisotopic mass and charge state of the precursor ion by analyzing a higher resolution MS1 “Zoom” scan that is acquired immediately before the MS2 scan of the precursor. The acquisition time of the MS1 Zoom scan is typically about half that required for the subsequent MS2 scan, reducing by about a third the number of MS2 scans acquired for a given period of time. However, we demonstrated that this reduction in the number of acquired scans did not reduce the number of peptide identifications in samples prepared from complex biological mixtures using the fractionation approaches described.33
The MS1 Zoom scans enable definitive determination of both the monoisotopic mass and charge state of the precursor ion by using MAZIE. As described in detail below, knowledge of the peptide charge and monoisotopic mass allows the PICquant algorithm to impose tighter tolerances when searching for the MS2 neutral mass losses and, thereby, enables a more accurate determination of the ratio of PIC-L to PIC-H labeled isotopomers for each individual MS2 scan, regardless of whether the peptide sequence can be deduced. While it is likely that the use of a high-resolution instrument for data acquisition would obviate the need for the extra MS1 Zoom scan, this data acquisition strategy enables the use of a low-resolution Thermo LTQ instrument, providing high precision mass and charge data of the precursor ion at a minimal cost of lost data.
PIC-Labeled Peptide Isotopomers Co-elute on Liquid Chromatography
Co-elution of isotopomer peptides during chromatography greatly simplifies comparison of labeled peptide abundance. Quantification experiments comparing the mass-tagged forms of non-coeluting isotopomers are at risk of overestimating the abundance of one tagged species at the beginning of an elution profile while underestimating the abundance near the end of the elution profile. Supplemental Figure 1 displays the ion chromatogram profiles of a representative isotopomer peptide pair that were not differentially expressed between the control and patient pools. The isotopomer pair exactly co-elutes during reverse phase chromatography as demonstrated by the consistency of the log2 of the calculated ratio between the ion intensities of the PIC-L and PIC-H derivatives. Thus accurate quantification of isotopomer ratios can be determined from each individual scan.
PIC-labeled peptides have unique neutral loss signatures
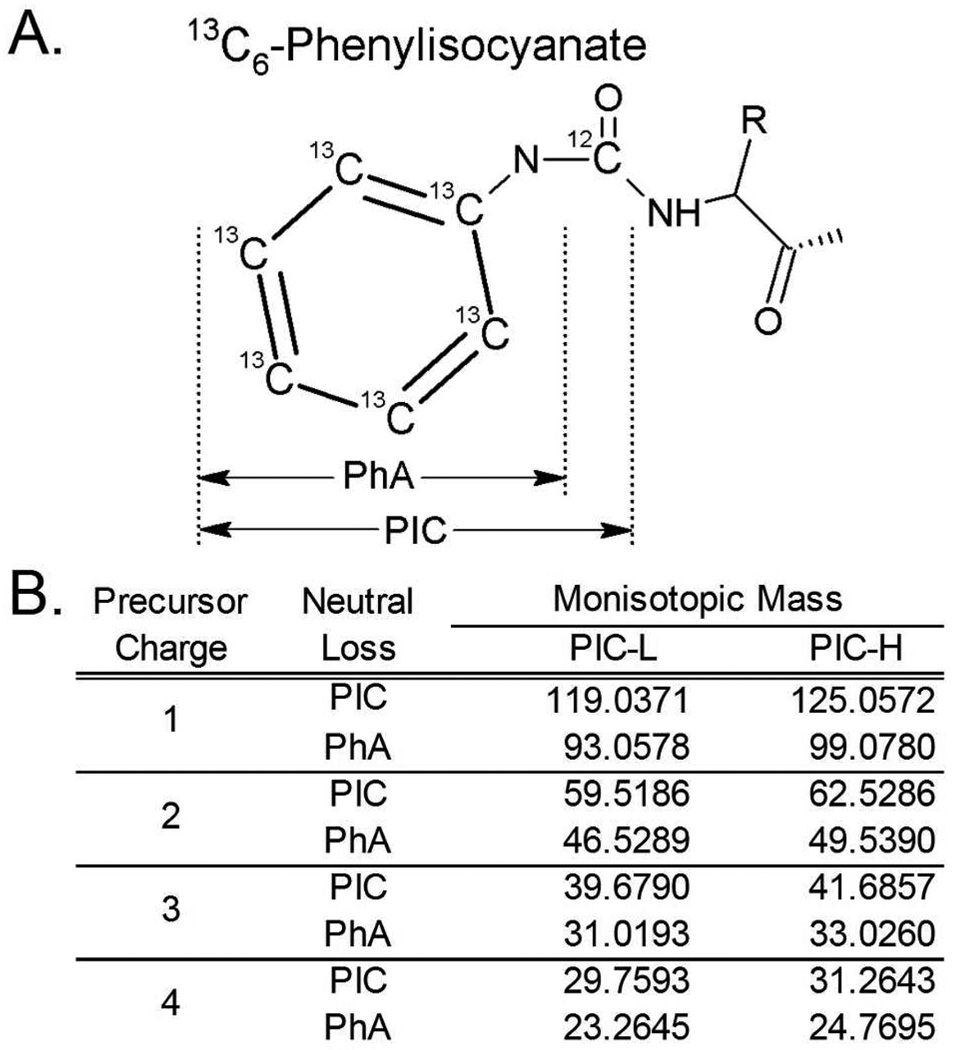
N-terminal labeling of peptides with PIC results in an asymmetrical urea bond between the phenyl and the peptide. MS2 scans generated by collision-induced dissociation (CID) fragmentation of PIC-labeled peptides include neutral mass losses resulting from cleavage across either side of the carbonyl, that correspond to loss of either the full phenylisocyanate (PIC) and the phenylamine (PhA) fragment, as illustrated in Figure 2A. These neutral mass losses are unique to the charge state and PIC label of the precursor ion (Figure 2B), and are also distinct from the product ions generated by any natural amino acids. The PICquant algorithm exploits this characteristic in a manner used previously for identifying neutral losses observed in phosphorylated peptides.39 Identification of these neutral mass losses in the MS2 scans of a data file confidently identifies PIC-labeled peptide ions.
Figure 2.
(A) The molecular locations of MS2 CID fragmentation of the asymmetrical urea bond that is associated with PIC labeling of peptide amino termini. Proximal cleavage results in the loss of a phenylamine (PhA) moiety from the peptide, while fragmentation at the distal location results in the loss of the entire PIC moiety. (B) The predicted neutral mass losses, as a function of the precursor charge state, for peptides labeled with either 12C6 (PIC-L) or 13C6 (PIC-H) phenylisocyanate when fragmented by CID.
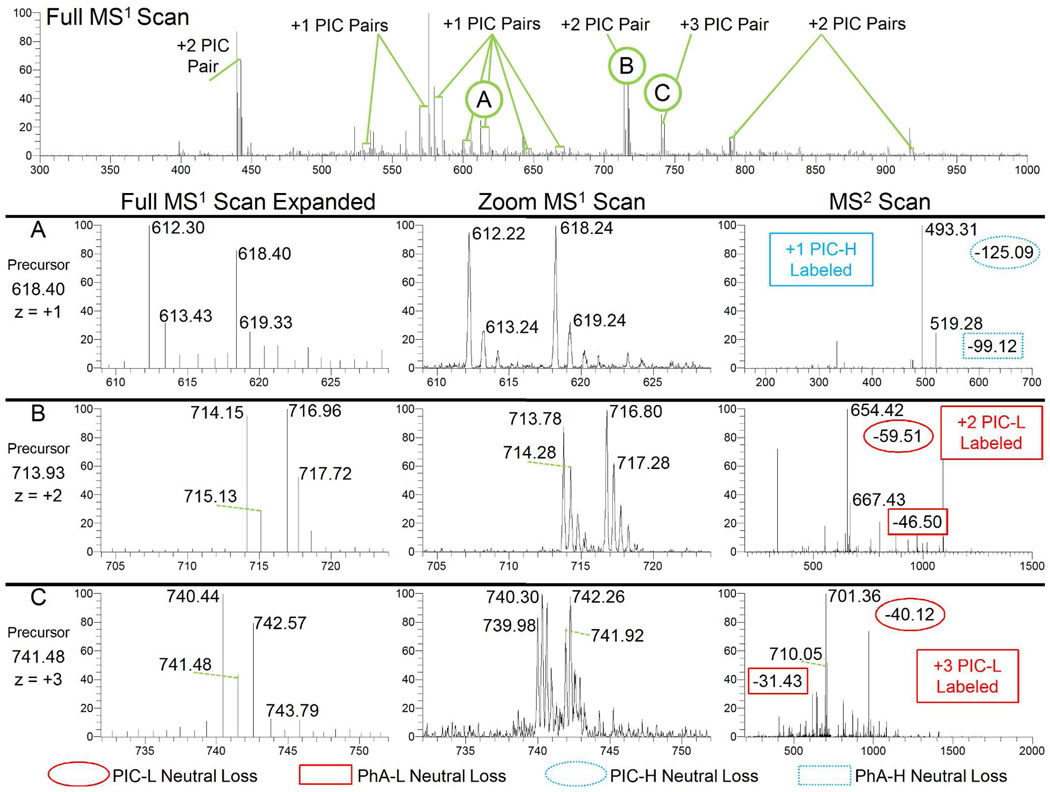
Example scans processed using PICquant are shown in Figure 3. The full-range MS1 scan (top) reveals numerous peptide ion pairs separated by 6, 3, or 2 Da, representing peptide isotopomer pairs from charge states +1, +2, and +3, respectively. Representative ion pairs of z = 1 to 3 are shown (panels A–C). Comparisons between conventional MS1 scans (left) and MS1 Zoom scans (middle) reveal the value of the Zoom scan, enabling quantification and accurate charge determination that is obscured in the lower resolution scan. In addition, the MS2 scans (right) illustrate the characteristic PhA neutral loss (box) and the PIC neutral loss (oval). These losses identify the 12C6- or 13C6-PIC label of an isotopomer peptide pair and confirm its precursor charge state.
Figure 3.
Representative data from a differentially PIC-labeled human urine tryptic digest. The top panel displays a full MS1 scan that contains multiple isotopomer peptide pairs with different charge states while Panels A–C illustrate some of these precursor ions that were subsequently selected for MS2 fragmentation. Each Panel A–C contains an expanded view of the original MS1 scan that matches the ±10 m/z range of the MS1 Zoom scan that was obtained immediately prior to the MS2 scan of the selected precursor. Within the MS2 scans, the ions associated with the PIC-H and PIC-L neutral mass losses are further labeled with respect to the original precursor mass to facilitate the use of the table in Figure 2B. In Panel A, for example, the two strongest peaks in the MS2 scan for the 618.40 m/z precursor correspond to neutral mass losses (618.40-493.31 = 125.09 and 618.40-519.28 = 99.12) that match those associated with a PIC-H label from a peptide with a +1 charge state. This charge state is confirmed in the preceding MS1 Zoom scan by both the 6 m/z separation between the two isotopic distributions observed and the 1 m/z spacing of ion peaks within each distribution. PICquant calculated a PIC-L/PIC-H ratio of 0.95 by taking the ratio between the intensities at m/z 612.22 and 618.24. Panels B and C illustrate similar examples for isotopomer peptide pairs with, respectively, +2 and +3 charge states. Note that the expanded view of the original MS1 scan illustrates that it lacks sufficient mass resolution.
The PICquant Algorithm
Numerous software packages for the quantification of isotope tags are available in both commercial and open source forms, most typically tailored for a specific tag, instrument, and/or methodology.4,20,40–43 Most rely upon quantifying only those peptides that can be confidently sequenced through the MS2 scans; some use isotopic labels that induce differential chromatographic mobility that must then be reconciled; and many are focused on determining ratios for proteins and assume that all matched peptides originate from the same protein and thus should have the same isotopic ratio.
To optimize the data analysis workflow and take full advantage of the unique features of the PIC label, we developed the PICquant algorithm. For each MS2 scan, PICquant considers the potential for either PIC-L or PIC-H labeling by predicting the two m/z ions that would be generated by the neutral mass losses, as tabulated in Figure 2B. For each charge state from +1 to +4, PICquant calculates a score based upon the product of the normalized MS2 ion intensity at these two m/z values for both PIC label types. Referring to Panel A of Figure 3 for a detailed example, the PIC and PhA neutral losses occur at 493.31 and 519.28, respectively, for the z = +1 ion of the PICH-labeled peptide; thus the score associated with the +1 charge state of the PIC-H label is 1.00*0.25 = 0.25.
The calculation of the PICquant score in this manner provides several advantages. First, demanding the existence of both neutral mass losses being present for each PIC-labeled peptide significantly reduces the likelihood that they represent peptide fragment ions that are completely unassociated with a PIC label. By also insisting that the charge state associated with the two PIC neutral losses is consistent with the charge state previously determined by the MAZIE algorithm, the probability of a false positive identification of a PIC label is significantly minimized.
Second, the product of the ion intensities provides a metric to further filter incidental ions that originate from the PIC-labeled peptide or a different peptide that elutes coincidentally. Though extensive sample fractionation significantly helps mitigate the frequency, complex biological samples will certainly have multiple ions with near identical elution and m/z properties. This could potentially distort a measured PIC-L/PIC-H ratio if the co-eluting ion is of comparable intensity to one of the PIC-labeled isotopomers. However, by demanding that the PIC score pass an empirically derived threshold, we ensure that the PIC-labeled isotopomer is indeed the dominant ion in a manner analogous to sequencing methodologies.
If PICquant determines that a MS2 scan contains a PIC-labeled peptide, the algorithm then examines the preceding MS1 Zoom scan. Combining the monoisotopic mass, charge state and PIC label identity determined by the MAZIE and PICquant algorithms, PICquant then identifies the peptide isotopomer pair and calculates the PIC-L/PIC-H ratio of the peak intensities associated with their monoisotopic masses. As with any sequence matching effort, false positive identifications and incorrect PIC-L/PIC-H ratio calculations are inevitable. To directly examine the effect of these issues, an experiment was conducted with a BSA standard sample that was labeled with a 1:1 PIC-L:PIC-H ratio and then processed normally. Fully detailed in Supplemental Materials, the narrow ln2(PIC-L/PIC-H) distribution of the standard sample had a mean of 0.12±0.86, demonstrating that the PICquant algorithm minimizes these identification errors and suggesting that PIC ratios that are significantly different from 1:1 in complex samples represent differentially expressed peptides that merit further validation.
Using PICquant to analyze each MS2 CID spectrum acquired from the 10 SDS-PAGE gel fractions and 11 IEF gel fractions, we identified 103,184 scans out of the total 293,843 MS2 scans (35%) as having either a PIC-L or PIC-H label. The remaining scans included those with low information content, non-peptidic ions, and peptides that had escaped PIC labeling. We applied a 2% False Discovery Rate (FDR) filter44 on the OMSSA sequencing results to generate a list of 17,576 scans with high-confidence peptide matches. From these scans, 1,030 unique peptides and 390 unique proteins were represented (see Supplemental Material), with roughly 70% of the peptides being identified with a PIC label.
Quantification of PIC-Labeled Peptide Isotopomers
As discussed above, we acquired a ±10 m/z MS1 Zoom scan before each MS2 scan in order to determine the accurate monoisotopic mass and charge state of the precursor peptide ion using MAZIE.33 The width of the Zoom scan was chosen to ensure that the full isotopic distribution of the isotopomer ion pair would also be observed within the scan, even for a +1 charge state for either a PIC-L or PIC-H labeled peptide. Because the PIC-labeled isotopomers exactly co-elute chromatographically, each Zoom scan can also be used to quantify the ratio of intensities for the monoisotopic mass of the peptide pair without having to sum intensities across multiple MS1 scans. Any additional scans acquired for the peptide (e.g. those with a different precursor charge state or PIC label) provide independent confirmation of the PIC-L/PIC-H ratio.
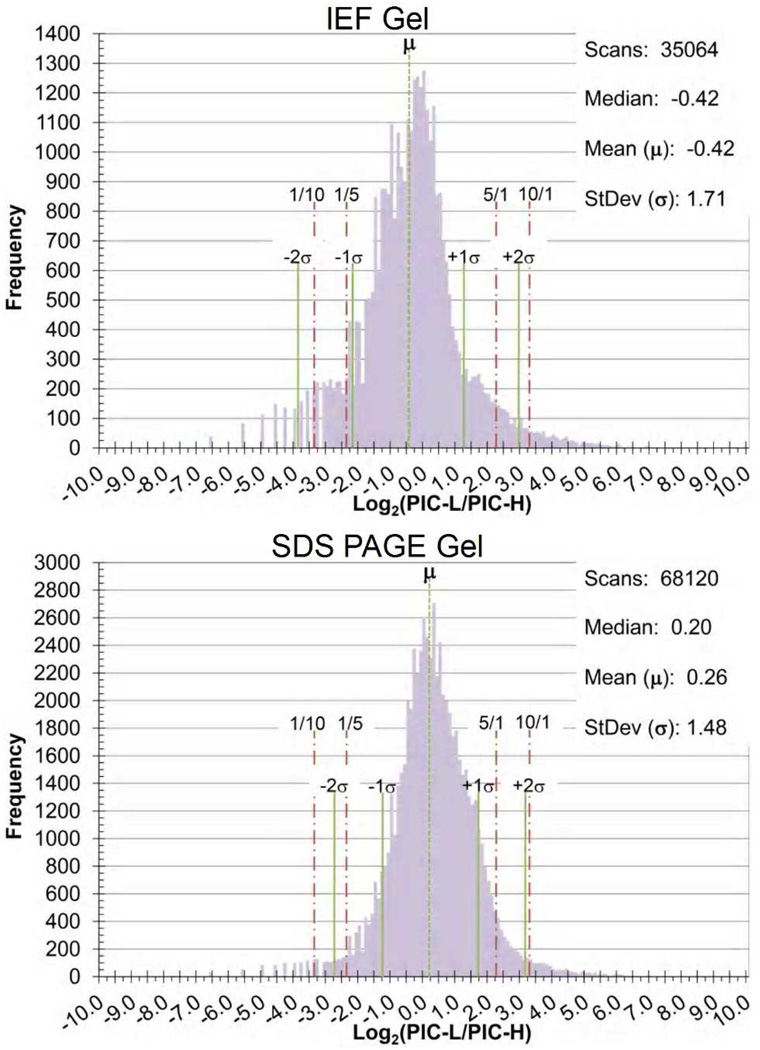
The histograms of Figure 4 present the log2 distribution of the PIC-L/PIC-H ratio for 35,064 and 68,120 mass tagged peptides from the IEF and SDS-PAGE gel fractions, respectively. We observed quantitative differences over a dynamic range of 104 in peptide concentration, with the vast majority having peak ratios near 1:1. The standard deviations of each histogram are 1.71 and 1.48 for the IEF fractions and SDS-PAGE fractions respectively, while the means were −0.42 and 0.26. Since the pooled patient sample for the two fractionation methods were inversely labeled, the mean and median of the distributions consistently indicate a slight bias towards a higher peptide concentrations in the patient sample.
Figure 4.
Histograms of the log2 of the PIC-L/PIC-H ratios calculated for results obtained from both the IEF peptide fractionation and the SDS-PAGE protein fractionation of PIC-labeled human urine. The histograms display the results obtained from the individual MS1 Zoom scans preceding the MS2 scan of the precursor ion. The standard deviations from its mean are marked by a solid line while the 1-to-10, 1-to-5, and their inverse ratios are marked by a dash-dot line. The granularity of the data at the low end of the histograms reflects the 0.01 resolution of the calculated PIC-L/PIC-H ratio.
“PRIDE” Grouping of Scans from Identical Peptide Precursors
To consolidate and merge the information derived from multiple sample fractions and data analyses, the calculations and information extracted from each MS2 scan from each fractionation technique were uploaded into two dedicated MySQL relational databases. Data storage was efficient, with the 18 and 19 mass spectral data files being summarized by a database of 18.6 and 21.4 megabytes for the IEF and SDS-PAGE fractionation, respectively. The database tables include the monoisotopic mass and charge state of the precursor ion as determined by the MAZIE algorithm; the peptide sequence and related information, if determined via the OMSSA search engine; and whether or not it represented a PIC-labeled isotopomer, as determined by the PICquant algorithm, along with the associated PIC-L/PIC-H ratio.
The MS2 scans from this database were grouped into collections designated Peptide Registry ID Elements (PRIDEs). Each peptide may exist with several charge states, each with a unique m/z, and each of these ions may be sampled multiple times. Furthermore, both the PIC-L and PIC-H isotopomer peptides may be sampled and quantified independently and some peptides will be found in multiple fractions. Grouping all of this data for each peptide into a single PRIDE so that the information can be analyzed collectively provides a means to maximize the quantitative value of these replicate scans. The PRIDE groups also greatly facilitate manual sequencing efforts by enabling the identification of the mass-tag-shifted b-ions upon comparing the MS2 scans of the isotopomer peptides. Thus, even without a confident sequence match, sufficient information for sequence determination can often be assembled for peptides of interest.
The grouping of the PRIDEs was done principally by comparing the accurate z = +1 monoisotopic “Root Mass” of the precursor of each MS2 scan. The Root Mass is the monoisotopic mass of the peptide before PIC labeling, calculated by subtracting the PIC label mass determined by PICquant from the accurate precursor monoisotopic mass identified by MAZIE. As a consequence, the Root Mass is the same for all ions analyzed for a particular peptide, regardless of charge or label, and is independent of sequence identification. Thus PRIDEs contain, in theory, all scans analyzed for a given peptide. Secondarily, a normalized chromatographic retention time associated with the precursor of the MS2 scan was used to help group scans into PRIDEs. Retention times for each sample were normalized by a spiked-in standard peptide that elutes near the start of the gradient and a polymer series that elutes near the end of the gradient. Determined empirically, the tolerances used for PRIDE grouping were 0.25 Da for the +1 monoisotopic mass and 10% for the normalized time, where the spiked-in peptide standard and the polymer series elution times marked the 0% and 100% normalized times, respectively. Histograms analogous to those in Figure 4 for these PRIDE groups are displayed in Supplemental Figure 3.
For a technical reproduciblilty analysis of the data set, the mass spectral acquisitions were acquired in duplicate for 7 IEF fractions and randomly organized into two groups. Desribed in detail in Supplemental Material, the PIC-L/PIC-H ratios for the 470 PRIDES with 5 or more scans had a Pearson’s r of 0.955, demonstrating strong reproducability.
Examination of Differentially Expressed Peptides with Sequence Identification
Even with high-resolution instruments, the majority of identified peptides have MS2 scans with a signal-to-noise (S/N) ratio of less than 10.45 Although low signal levels of the preceding MS1 Zoom scan does not necessarily correlate with low quality MS2 fragmentation, it does limit the PIC-L/PIC-H ratio that can be calculated. We anticipate that the differentially expressed proteins/peptides will most frequently be in low abundance. Hence, they would have relatively low S/N MS1 intensity levels and the calculated PIC-L/PIC-H ratios of the potential biomarker candidates will most likely be on the order of 10 (or conversely, 1/10), roughly corresponding to two standard deviations (±2σ) from the distribution mean of the sample (Figure 4).
We generated a candidate list of differentially expressed peptides by querying the MySQL relational database for these PIC-L/PIC-H ratio extremes. The majority of PRIDES with PIC ratios that are significantly different from the sample mean represented peptides of unknown sequence. While additional methods such as de novo sequencing can be applied to these scans, we proceeded to validate our protocol by examining those scans that also had a high-confidence sequence identification (Supplemental Table 3). Both the peptide sequence and the PIC-L/PIC-H ratio associated with the PRIDE were manually validated. Because the uncertainty of the calculated PIC ratio decreases with higher S/N levels in the scan, we weighted the PIC ratios measured in individual Zoom scans by the ion intensity of the monoisotopic mass, and then calculated mean and standard deviation for measurements in each PRIDE.
As seen in Supplemental Table 3, the number of scans identified for each peptide ranged from a high of 62 to a low of 1 individual scan. These represent the inclusion of all scans for each of the different charge states and PIC labels that were collected into a particular PRIDE. Three distinct peptides were identified from each of five proteins, and two distinct peptides were identified from each of another five proteins. Examining one of these proteins in detail, S100 protein A9 (gi 4506773) was identified from three peptides, representing 51 individual scans, which had a mean PIC ratio of less than 1/10. The value of being able to rapidly identify proteins with numerous confirmatory quantifications is evident. Moreover, PRIDEs that have numerous replicate quantifications rapidly point to scans that merit additional effort to determine sequence identity.
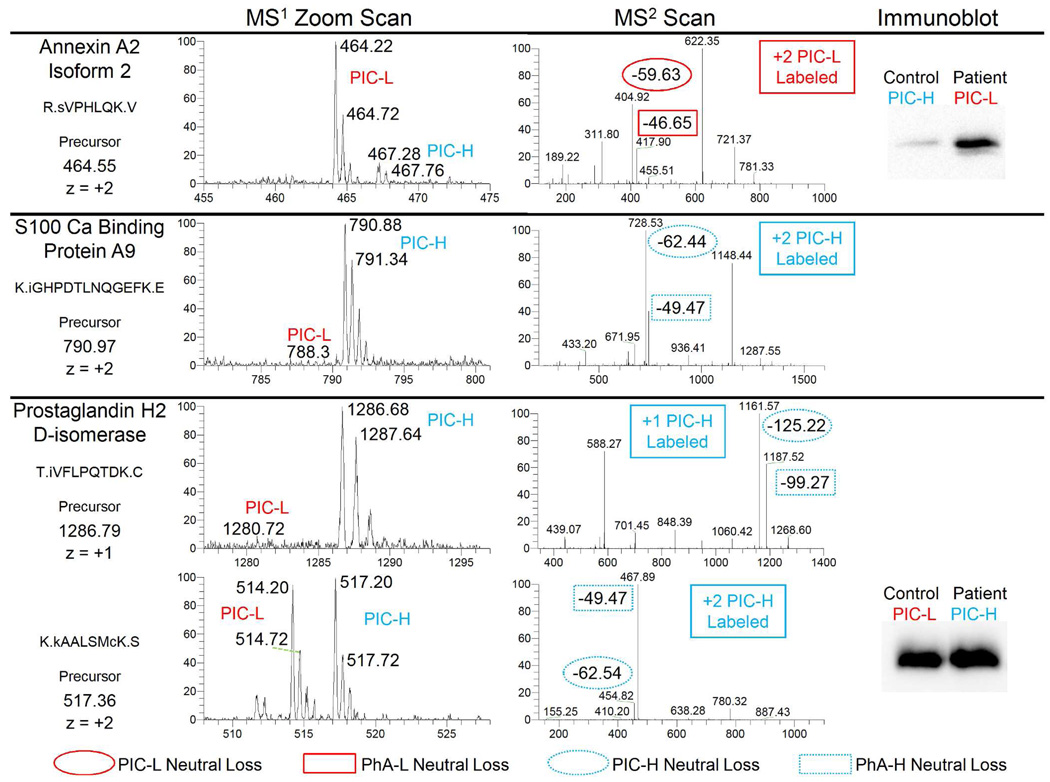
Figure 5 illustrates representative scans that were acquired for several of these candidates. Comparing the MS1 Zoom scans of these candidates with the scans displayed in Figure 3 clearly illustrates the differential expression of these candidate peptides between the pooled patient and control samples. Note in particular that the MS1 Zoom scan that is associated with the 790.97 precursor essentially contains an ion singlet (middle panel). Using other mass-tag strategies, if no sequencing result was obtained, it would be impossible to determine if this ion represented a differentially expressed peptide or was derived from an unlabeled form of a peptide or an extraneous contaminant. However, the unique PIC neutral mass losses of the following MS2 scan enable the ion to be identified confidently as a PIC-H labeled peptide of interest. The immunoblot for the annexin A2 protein (top panel of Figure 5) provides independent confirmation of its significant differential expression in the pooled samples.
Figure 5.
Representative scans from a few of the differentially expressed peptides identified from urine that was obtained from breast cancer patients. The MS1 Zoom scans illustrate the intensities from which the corresponding PIC ratios listed in Supplemental Table 3 were calculated. The neutral mass losses in the MS2 scans are labeled in a manner similar to Figure 3. The immunoblots support the observed peptide quantitation identified by the PICquant platform via mass spectrometry. Note that the prostaglandin H2 D-isomerase semi-tryptic peptide T.IVFLPQTDK.C is C-terminal to the epitope of the antibody while K.kAALSMcK.S is at the epitope.
Evidence of differential post-translational modifications of proteins was also detected. The bottom panel of Figure 5 displays the results for two different peptides of prostaglandin H2 D-isomerase. The K.kAALSMcK.S peptide and at least 4 others (Supplemental Figure 6), have PIC ratios near 1. The displayed immunoblot, with an epitope flanked by these peptides, supports this result. However, the semi-tryptic peptides T.IVFLPQTDK.C (shown) and D.TIVFLPQTDK.C (Supplemental Table 3) are C-terminal to these other peptides and were observed a total of 82 times. Both display a strong differential expression between the patient and control samples. This discrepancy in their PIC-L/PIC-H ratios could be explained by disease-relevant proteolytic activity or by the differential expression of one of the many observed splice variants of this protein. We were unable to obtain antibodies that probed this region of the protein for confirmation. However, using the PICquant strategy alone, this post-translational modification of the protein was quantified successfully, illustrating the advantage of examining the samples at the peptide level as opposed to just at the protein level.
While we focused this effort on those differentially-expressed peptides for which high confidence sequence matches were available, it is important to note that the immediate sequencing of the candidate peptide ions is not essential to the success of the PICquant platform. Once a set of peptide ions has been identified as being potential biomarkers, inclusion lists can be generated and coupled with multiple reaction monitoring (MRM) mass spectrometry. This approach is already established in label-free biomarker identification efforts.29 The advantage of MRM mass spectrometry is that it can isolate peptides at relatively low abundance in complex clinical samples without the need for laborious fractionation. This approach provides two important advantages. First, MS2 fragmentation data can be readily obtained for the specific peptide ions of interest without the PIC isotope mass tag, increasing the probability of successfully sequencing the peptide candidates. Second, by allowing examination of individual instead of pooled samples, a preliminary investigation into the clinical viability of the potential biomarkers can be undertaken in a relatively high-through-put manner. A clinically relevant biomarker must be relatively insensitive to the proteomic variability among humans. A demonstration of the MRM mass spectrometry approach to evaluate the initial clinical viability of the potential biomarkers identified using the PICquant platform is currently ongoing.
We emphasize that the differentially expressed proteins/peptides described here should not be interpreted as potential markers of cancer. We have used these samples only as representatives of typical clinical samples for this proof of concept study, and have not demonstrated that the differentially detected peptides represent disease correlates. We do not anticipate that this protocol would be used in a clinical setting due to the increased analysis time and efforts required. Instead, the goal of the PICquant platform is to identify candidate peptide biomarkers. Antibodies to these candidates can then be used to rapidly analyze the large number of clinical samples necessary to establish diagnostic specificity and sensitivity.
Conclusions
We have developed a protocol using 12C6- and 13C6-phenylisocyanate to quantify differential peptide abundance in complex biological samples for candidate protein biomarker discovery. This PICquant platform combines non-novel, individual components into an automated protocol that represents a unique approach. First, because of the unique neutral mass losses of PIC-labeled peptides undergoing CID fragmentation, it can identify and quantify differentially expressed peptides independent of sequence determination. Second, the exact co-elution of PIC-labeled isotopomers enables quantification that is independent of chromatographic variabilities and enables statistically relevant quantification even if the peptide was selected for MS2 fragmentation only once. Third, the grouping of the peptide isotopomer pairs into PRIDEs quickly simplifies complex data sets, improves the statistical relevance of the quantification, and facilitates subsequent targeted sequencing efforts of potential peptide candidates. Thus, the PICquant platform represents a valuable new approach to peptide quantification that is efficient, relatively inexpensive and applicable to a broad range of biological and clinical samples.
Supplementary Material
Acknowledgments
The authors thank Meera Murgai, Fang Xu, and Michael Kidd for their assistance in collecting, labeling, and storing the urine samples from the breast cancer patients. We also thank Fang Xu for her assistance in conducting the immunoblots and Meera Murgai for her assistance with the development of the MySQL database and its interface. Funding for this work was provided through the National Cancer Institute, grant number CA126101.
Footnotes
Supporting Information Available: Supporting information also includes a more detailed description of sample preparation; mass spectral acquisition and analysis; an evaluation of PIC-labeling; a charge state break-down of the peak areas for the PIC-labeling of the BSA digest test sample in Supplemental Table 1; PIC-labeled peptide isotopomer co-elution in Supplemental Figure 1; an analysis of a BSA tryptic digest labeled 1:1 with PIC-L:PIC-H in Supplemental Figure 2; a histogram of the average log2(PIC-L/PIC-H) ratio calculated from all the PIC-labeled scans grouped into a PRIDE in Supplemental Figure 3; a technical reproducibility analysis in Supplemental Figure 4; a charge state break-down of identified MS2 scans in Supplemental Table 2; a list of differentially expressed peptides identified in the urine samples in Supplemental Table 3; annotated MS2 mass spectra for the peptides in Supplemental Table 3 with relatively poor OMSSA E-values in Supplemental Figure 5; an annotated sequence of prostaglandin H2 D-isomerase in Supplemental Figure 6; and a display of the PRIDEs with respect to their average monoisotopic mass and normalized retention time for the IEF and SDS-PAGE fractions in Supplemental Figure 7 and 8, respectively. Also included is a spreadsheet tabulating the identified peptides that passed a 2% FDR filter of the OMSSA search results for both fractionation techniques. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Goshe MB, Smith RD. Current Opinion in Biotechnology. 2003;14:101–109. doi: 10.1016/s0958-1669(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 2.Tao WA, Aebersold R. Current Opinion in Biotechnology. 2003;14:110–118. doi: 10.1016/s0958-1669(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 3.Cox J, Mann M. Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 4.Matthiesen R, Carvalho AS. In: Bioinformatics Methods in Clinical Research. Matthiesen R, editor. Vol. 593. Humana Press; 2010. pp. 187–204. [Google Scholar]
- 5.Smolka MB, Zhou HL, Purkayastha S, Aebersold R. Analytical Biochemistry. 2001;297:25–31. doi: 10.1006/abio.2001.5318. [DOI] [PubMed] [Google Scholar]
- 6.Gygi SP, Rist B, Griffin TJ, Eng J, Aebersold R. Journal of Proteome Research. 2002;1:47–54. doi: 10.1021/pr015509n. [DOI] [PubMed] [Google Scholar]
- 7.Ranish JA, Yi EC, Leslie DM, Purvine SO, Goodlett DR, Eng J, Aebersold R. Nature Genetics. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 8.Goodlett DR, Keller A, Watts JD, Newitt R, Yi EC, Purvine S, Eng JK, von Haller P, Aebersold R, Kolker E. Rapid Communications in Mass Spectrometry. 2001;15:1214–1221. doi: 10.1002/rcm.362. [DOI] [PubMed] [Google Scholar]
- 9.Hood BL, Lucas DA, Kim G, Chan KC, Blonder J, Issaq HJ, Veenstra TD, Conrads TP, Pollet I, Karsan A. Journal of the American Society for Mass Spectrometry. 2005;16:1221–1230. doi: 10.1016/j.jasms.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Sakai J, Kojima S, Yanagi K, Kanaoka M. Proteomics. 2005;5:16–23. doi: 10.1002/pmic.200300885. [DOI] [PubMed] [Google Scholar]
- 11.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Molecular & Cellular Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Koehler CJ, Strozynski M, Kozielski F, Treumann A, Thiede B. Journal of Proteome Research. 2009;8:4333–4341. doi: 10.1021/pr900425n. [DOI] [PubMed] [Google Scholar]
- 13.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Molecular & Cellular Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Hinsby AM, Olsen JV, Mann M. Journal of Biological Chemistry. 2004;279:46438–46447. doi: 10.1074/jbc.M404537200. [DOI] [PubMed] [Google Scholar]
- 15.Ong SE, Mann M. Nature Protocols. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 16.Hanke S, Besir H, Oesterhelt D, Mann M. Journal of Proteome Research. 2008;7:1118–1130. doi: 10.1021/pr7007175. [DOI] [PubMed] [Google Scholar]
- 17.Elias JE, Haas W, Faherty BK, Gygi SP. Nature Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 18.Wong JW, Sullivan MJ, Cartwright HM, Cagney G. BMC Bioinformatics. 2007;8:51. doi: 10.1186/1471-2105-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubner NC, Ren S, Mann M. Proteomics. 2008;8:4862–4872. doi: 10.1002/pmic.200800351. [DOI] [PubMed] [Google Scholar]
- 20.Mueller LN, Brusniak MY, Mani DR, Aebersold R. Journal of Proteome Research. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 21.Wong JW, Cagney G. In: Proteome Bioinformatics. Hubbard SJ, Jones AR, editors. Vol. 604. Humana Press; 2010. pp. 273–283. [Google Scholar]
- 22.Listgarten J, Emili A. Molecular & Cellular Proteomics. 2005;4:419–434. doi: 10.1074/mcp.R500005-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Theodorescu D, Wittke S, Ross MM, Walden M, Conaway M, Just I, Mischak H, Frierson HF. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- 24.Letarte S, Brusniak MY, Campbell D, Eddes JS, Kemp CJ, Lau H, Mueller LN, Schmidt A, Shannon P, Kelly-Spratt KS, Vitek O, Zhang H, Aebersold R, Watts JD. Clinical Proteomics. 2008;4:105–116. doi: 10.1007/s12014-008-9018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finney GL, Blackler AR, Hoopmann MR, Canterbury JD, Wu CC, MacCoss MJ. Analytical Chemistry. 2008;80:961–971. doi: 10.1021/ac701649e. [DOI] [PubMed] [Google Scholar]
- 26.Liu HB, Sadygov RG, Yates JR. Analytical Chemistry. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 27.Zybailov B, Coleman MK, Florens L, Washburn MP. Analytical Chemistry. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 28.Hendrickson EL, Xia QW, Wang TS, Leigh JA, Hackett M. Analyst. 2006;131:1335–1341. doi: 10.1039/b610957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteaker JR, Zhang H, Zhao L, Wang P, Kelly-Spratt KS, Ivey RG, Piening BD, Feng LC, Kasarda E, Gurley KE, Eng JK, Chodosh LA, Kemp CJ, McIntosh MW, Paulovich AG. Journal of Proteome Research. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 30.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Molecular & Cellular Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Usaite R, Wohlschlegel J, Venable JD, Park SK, Nielsen J, Olsson L, Yates JR. Journal of Proteome Research. 2008;7:266–275. doi: 10.1021/pr700580m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shevchenko A, Wilm M, Vorm O, Mann M. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 33.Victor KG, Murgai M, Lyons CE, Templeton TA, Moshnikov SA, Templeton DJ. Journal of the American Society for Mass Spectrometry. 2010;21:80–87. doi: 10.1016/j.jasms.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang XY, Shi WY, Bryant SH. Journal of Proteome Research. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 35.Regnier FE, Julka S. Proteomics. 2006;6:3968–3979. doi: 10.1002/pmic.200500553. [DOI] [PubMed] [Google Scholar]
- 36.Stark GR. Biochemistry. 1965;4:1030–1036. doi: 10.1021/bi00882a008. [DOI] [PubMed] [Google Scholar]
- 37.Mason DE, Liebler DC. Journal of Proteome Research. 2003;2:265–272. doi: 10.1021/pr0255856. [DOI] [PubMed] [Google Scholar]
- 38.Zhang RJ, Sioma CS, Thompson RA, Xiong L, Regnier FE. Analytical Chemistry. 2002;74:3662–3669. doi: 10.1021/ac025614w. [DOI] [PubMed] [Google Scholar]
- 39.Zarling AL, Ficarro SB, White FM, Shabanowitz J, Hunt DF, Engelhard VH. Journal of Experimental Medicine. 2000;192:1755–1762. doi: 10.1084/jem.192.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li XJ, Zhang H, Ranish JA, Aebersold R. Analytical Chemistry. 2003;75:6648–6657. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 41.MacCoss MJ, Wu CC, Liu H, Sadygov R, Yates JR., 3rd Analytical Chemistry. 2003;75:6912–6921. doi: 10.1021/ac034790h. [DOI] [PubMed] [Google Scholar]
- 42.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Nature Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 43.Mortensen P, Gouw JW, Olsen JV, Ong SE, Rigbolt KT, Bunkenborg J, Cox J, Foster LJ, Heck AJ, Blagoev B, Andersen JS, Mann M. Journal of Proteome Research. 2010;9:393–403. doi: 10.1021/pr900721e. [DOI] [PubMed] [Google Scholar]
- 44.Kall L, Storey JD, MacCoss MJ, Noble WS. Journal of Proteome Research. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- 45.Bakalarski CE, Elias JE, Villen J, Haas W, Gerber SA, Everley PA, Gygi SP. Journal of Proteome Research. 2008;7:4756–4765. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.