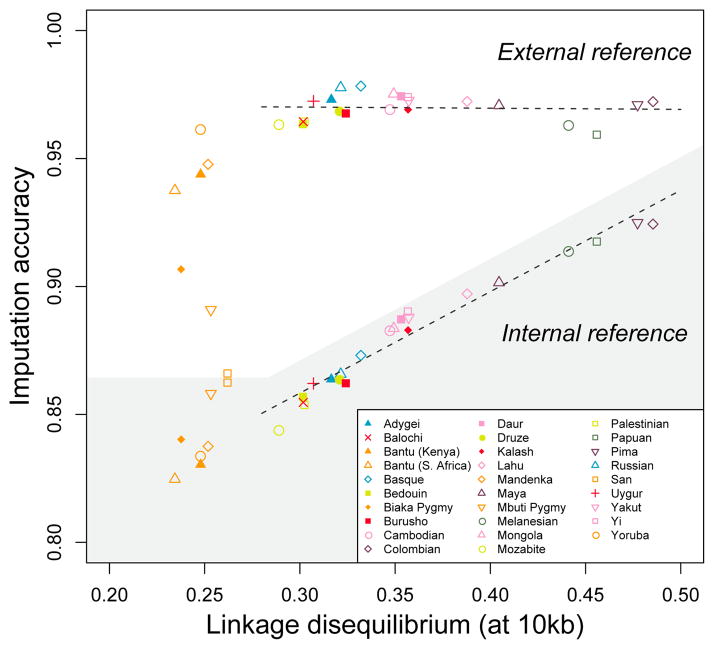
Figure 4. Genotype imputation accuracy in 29 populations, with and without external reference panels.
Imputation accuracy is plotted as a function of LD measured by mean r2 at a distance of 10 kb in a genome-wide dataset43. Genotypes in a genome-wide study are hidden and then imputed, with two different designs. In the shaded region, genotypes in each population are imputed without an external reference panel, so that the information for imputing “missing” genotypes comes from other individuals in the population. In the unshaded region, genotypes in the population are imputed using an external reference panel, chosen optimally among 36 mixtures of the HapMap CEU (European American), CHB+JPT (Chinese and Japanese), and YRI (Yoruba) panels. Color coding for populations follows that of Fig. 3. The regression lines exclude the African populations, and they have coefficients of determination 0.003 (external reference) and 0.953 (internal reference). The figure shows that imputation accuracy based on an internal reference is highly correlated with LD. However, imputation accuracy based on an external reference is not correlated with LD (and instead depends on the composition of the particular reference panels available). The figure is based on the data in scenarios 1, 3, and 6 in Table 1 of Huang et al. 68.