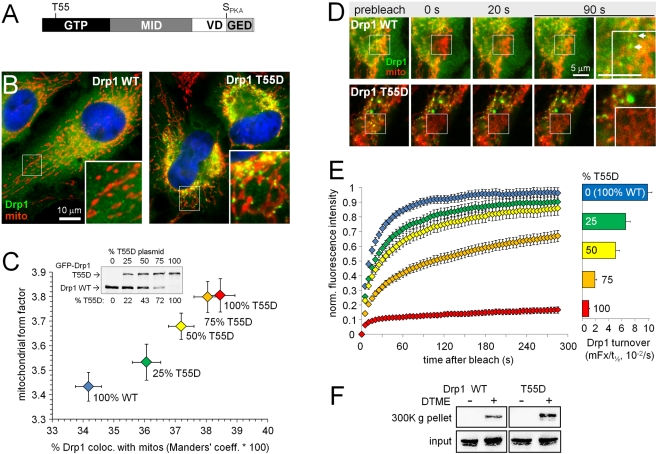
Figure 9. A GTPase-compromised Drp1 mutant accumulates in slowly recycling complexes at mitochondria.
(A) Drp1 domain diagram. (B–C) Endogenous Drp1 in HeLa cells was replaced by transfection of the indicated ratios of wild-type (WT) and T55D mutant GFP-Drp1 expression plasmids. Cells were fixed and analyzed for mitochondrial length and Drp1 colocalization with mitochondria using ImageJ software [22],[61]. (B) Representative images showing punctate localization of GFP-Drp1 T55D (green) on mitochondria (cytochrome oxidase subunit II antibody). (C) Correlation between mitochondrial length and mitochondrial localization of Drp1 as a function of Drp1 T55D expression (means ± s.e.m. of ∼200 cells per condition from a representative experiment). Wild-type and T55D-mutant Drp1 expression levels are similar and scale with plasmid amounts (inset). (D, E) Turnover of GFP-Drp1 WT and T55D was analyzed by FRAP as in Figure 8. (D) Frames showing representative time lapse series (green: GFP-Drp1, red: MitoTracker Deep Red, see Video S2) of HeLa cells expressing 100% WT or T55D Drp1, with the last frame expanded to show recovery of WT Drp1 into mitochondrial foci (arrows). (E) Average recovery curves (left) and curve fit-derived turnover (right, ratio of mobile fraction [mFx] and 50% recovery time [t1/2]) from cells expressing varying ratios of WT and T55D-mutant GFP-Drp1 (mean ± s.e.m. of 12–20 cells each from a representative experiment). (F) COS cells expressing GFP-Drp1 WT or T55D were incubated with DTME (5 min, 500 µM), and cleared cell lysates were subjected to ultracentrifugation to sediment Drp1 complexes (∼2-fold increase with the T55D mutation).