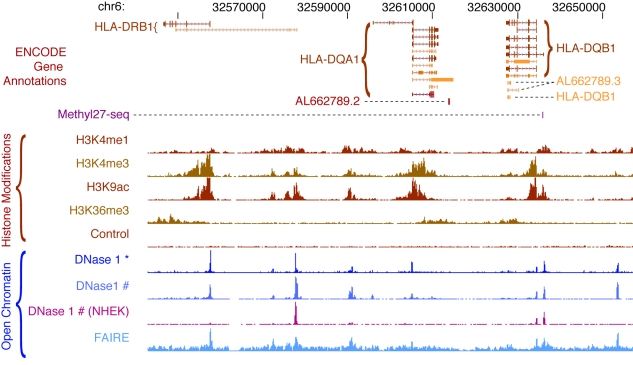
Figure 4. ENCODE chromatin annotations in the HLA locus.
Chromatin features in a human lymphoblastoid cell line, GM12878, are displayed for a 114 kb region in the HLA locus. The top track shows the structures of the annotated isoforms of the HLA-DRB1, HLA-DQA1, and HLA-DQB1 genes from the ENCODE Gene Annotations (GENCODE), revealing complex patterns of alternative splicing and several non-protein-coding transcripts overlapping the protein-coding transcripts. The purple mark on the next line shows that a CpG in the promoter of the HLA-DQB1 gene is partially methylated (assayed on the Illumina Methylation27 BeadArray platform). The densities of four histone modifications associated with transcriptionally active loci are plotted next, along with the input control signal (generated by sequencing an aliquot of the sheared chromatin for which no immunoprecipitation was performed). The last lines plot the accessibility of DNA in chromatin to nucleases (DNaseI) and reduced coverage by nucleosomes (FAIRE); peaks on these lines are DNaseI hypersensitive sites. Note that the ENCODE Consortium generates DNaseI accessibility data by two alternative protocols marked by * and #. The magenta track shows DNaseI sensitivity in a different cell line, NHEK, for comparison.