Figure 2.
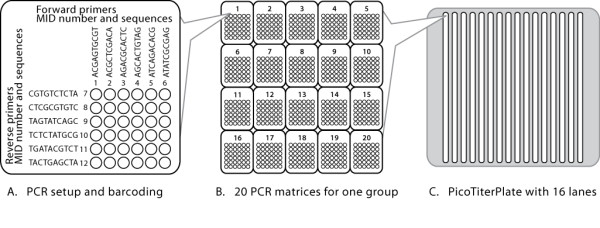
The experimental setup. In order to sequence 20 mitochondrial amplicons from 576 samples of Atlantic salmon, brown trout and Arctic charr, in a single run, the isolated DNA was divided into 16 groups, each containing 36 samples. The mitochondrial fragments were amplified from each group in a matrix of PCR reactions, using barcoded primers (MIDs) (A). 20 matrices of PCR reactions, one for each mitochondrion fragment, were carried out (B). Up to 720 amplicons from each group were pooled together in near equimolar concentrations. The 16 pools created were used to generate single stranded fragment DNA templates for the FLX sequencing. 16 amplified sst libraries, consisting of DNA fragments on beads, were loaded onto a PicoTiterPlate equipped with a sixteen-lane gasket where each library was assigned one lane (C). Based on the combination of MIDs on both sites of the amplicons within each group (library), individual sequencing reads were assigned to the corresponding samples.
