Figure 4.
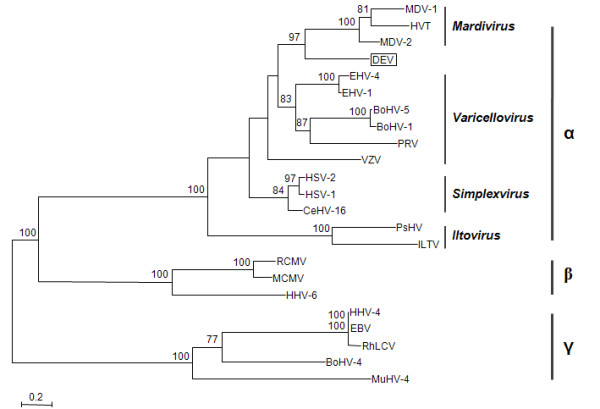
Phylogenetic tree based on the UL15 protein sequences of 23 herpesviruses. Amino acid sequences of UL15 homologues from 23 herpesviruses were retrieved and aligned using the ClustalW2 program. An unrooted phylogram was constructed using the PhyML software (version 3.0) with the LG amino acid substitution model and the non-parametric bootstrap support test. SeaView 4.1 was used for displaying and editing phylogenetic trees [43]. Values of bootstrap supports (100 replicates) in excess of 70% are shown on the shoulder of branches. The scale bar indicates 0.1 substitution/site and vertical lines are used to represent different subfamilies within the Herpesviridae and genera within the Alphaherpesvirinae.
