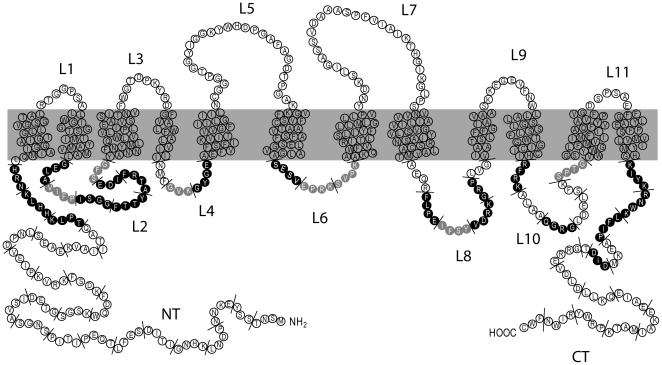
Figure 1. Schematic topology model of the Gap1 protein.
Residues shown in black or gray in the N-terminal tail (NT), C-terminal tail (CT) and intracellular loops (L2, L4, L6, L8, L10) are those which, when replaced with alanines, cause Gap1 to be inactive or partially active, respectively (see text). The short lines delineate the mutagenized blocks of 3 or 4 amino acids. The position of the ubiquitin-acceptor lysines (K9 and K16) and of specific, more deeply analyzed, mutations is shown.