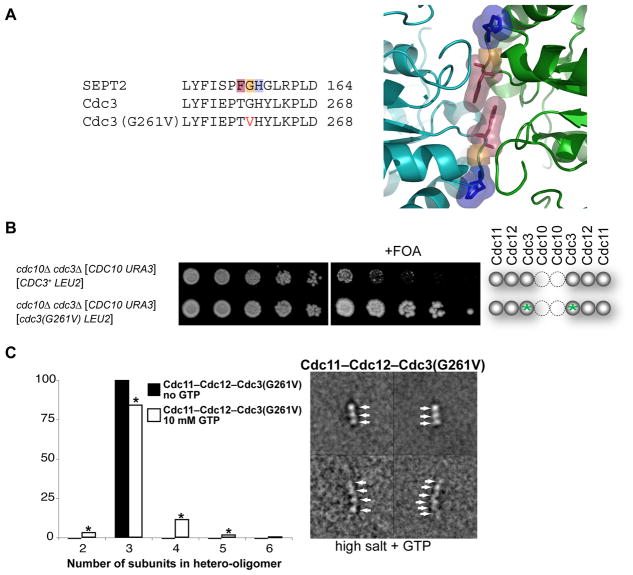
Figure 4. Mutation at the G Interface of Cdc3 Enhances Survival of Cdc10-less Cells.
(A) Sequence alignment (left) of human SEPT2 with WT Cdc3 and Cdc3(G261V). Highlighting (Phe, pink; Gly, orange; His, blue) refers to same residues shown (right) in the close-up of the G homodimer interface of GppNHp-bound mouse SEPT2 (PDB accession no. 3FTQ) (Sirajuddin et al., 2009). (B) As in 1C, except grown on SCGlc-Leu and incubated for 3 (left) or 5 dy (right); green asterisks, cdc3(G261V) mutation. (C) Purified recombinant Cdc11–Cdc12–Cdc3(G261V) complexes were examined by EM in 300 mM salt buffer in the absence (solid bars) and presence (open bars) of 10 mM GTP; frequencies of particle classes observed (representative examples on the right) plotted as histograms (left). All 824 particles (100%) in the absence of GTP were trimers; 3358 particles in the presence of GTP were 97 dimers (3%), 2831 trimers (84%), 376 tetramers (11%), 46 pentamers (1%), and 8 hexamers (0.2%). Asterisk, classes whose appearance depended on presence of GTP at a statistically significant level (P < 0.0001, according to a two-tailed Fisher’s exact test).