Figure 1. NG1427 is a transcriptional repressor that shares similarity to E. coli LexA.
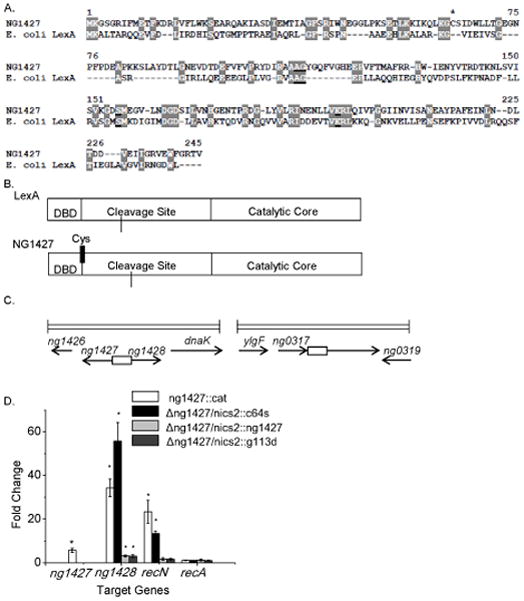
A: Amino acid sequence alignment comparing NG1427 and E. coli LexA. S24 peptidase cleavage and active sites are underlined and conserved residues are highlighted. Asterisk denotes single cysteine residue found in NG1427. B: Cartoon depiction of NG1427 and LexA protein domains. “DBD,” refers to the DNA binding domain. Location of single cysteine (Cys) in NG1427 is marked with black line. C: Diagram of the ng1427/ng1428 and recN chromosomal loci in FA1090 Gc. Boxes denote intergenic regions containing putative promoters of ng1427/ng1428 or recN. D: Fold change of target gene expression in ng1427::cat, Δng1427/nics2::c64s, Δng1427/nics2::g113d, and Δng1427/nics2::ng1427, relative to the parental strain as measured by qPCR with omp3 used as a reference gene. Asterisks denote p<0.05 as calculated by Student’s t-test relative to negative control (recA). Error bars +/− s.e.m. All data shown are the result of at least 3 biological replicates (n=3).
