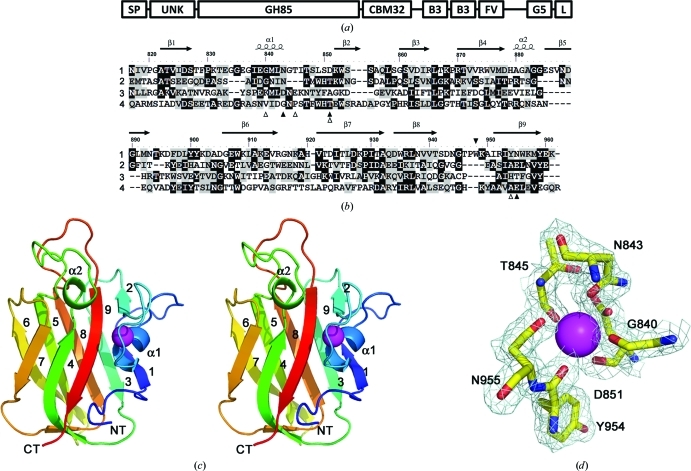
Figure 1.
The architecture of EndoD from S. pneumoniae (Sp_0498; 183.2 kDa) and the structure of SpCBM32. (a) The modular arrangement of EndoD. The enzyme is secreted to the extracellular surface, where its C-terminus becomes (L = LPXTG) covalently attached to the peptidoglycan in the cell wall through a sortase-mediated reaction. The catalytic GH85 module (172–816) is flanked by an N-terminal unknown domain (UNK, 38–170) and the SpCBM32 module (CBM32, 817–945), which lies towards the C-terminus. Several other modules are also predicted in the extensive C-terminal region of the molecule, including tandem bacterial Ig-like or Big3 domains (B3, 1067–1139, 1157–1225) and a FIVAR motif (FV, 1243–1287), which are commonly observed within bacterial surface proteins. The function of these C-terminal modules remains unknown. (b) Primary and secondary structure of SpCBM32 (1) in comparison with known structures: C. perfringens exo-α-sialidase appended CBM32 (2; PDB entry 2v72), B. thetaiotaomicron α-l-fucosidase appended CBM32 (3; PDB entry 3eyp) and M. viridifaciens sialidase CBM32 (4). The black triangles indicate the location of the unqiue Trp948 residue in SpCBM32 (above; filled) and the amino acids involved in calcium coordination (below; filled, side chain; open, main-chain carbonyl). (c) The three-dimensional structure of SpCBM32 in wall-eyed stereo cartoon format with a blue (N-terminal) to red (C-terminal) colour gradient and the structural calcium shown as a magenta sphere. The nine β-strands and two α-helices are labelled. (d) The calcium-binding site of SpCBM32. The pentagonal bipyramidal geometry of the desolvated coordination pocket is displayed with the weighted maximum-likelihood (Murshudov et al., 1997 ▶)/σA (Read, 1986 ▶) 2F obs − F calc map (cyan) contoured at 1.0σ (0.35 e− Å−3).