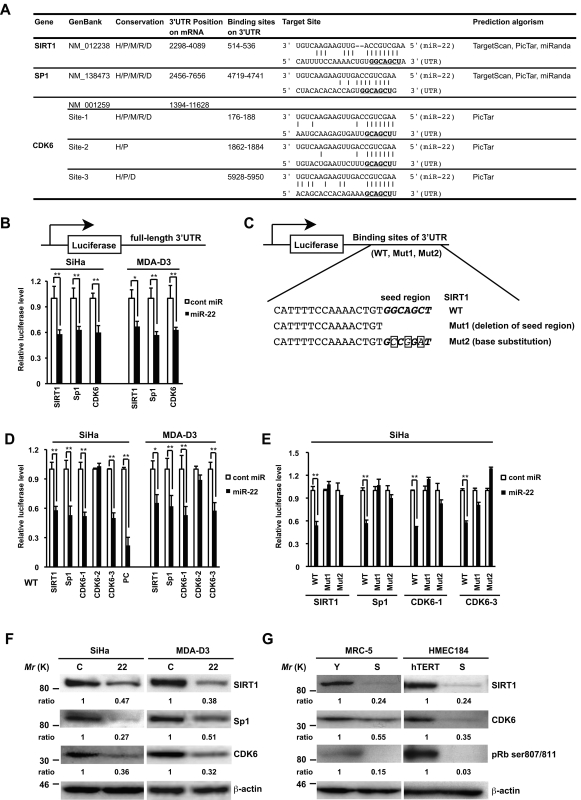
Figure 5.
SIRT1, Sp1, and CDK6 are direct targets of miR-22. (A) Summary of miR-22 target sites in the 3′-UTR of SIRT1, Sp1, and CDK6. H, human; P, chimp; M, mouse; R, rat; D, dog. The underlined bold nucleotides indicate target sites. (B–E) Target validation of SIRT1, Sp1, and CDK6 was confirmed in the luciferase reporter assay. (C) Scheme of the luciferase reporter constructs containing conserved miR-22 target sites (WT), deletion of seed region (Mut1), or base substitution of seed region (Mut2) was indicated using SIRT1 as an example. The seed region is italic bold, and point mutations are boxed in the seed region. Each construct, including full-length 3′-UTR (B), WT (D), or Muts (E), was cotransfected with miR-22 or cont miR into SiHa or MDA-D3 cells. Relative luciferase level = (Sluc/Srenilla)/(Cluc/Crenilla) in relative light units (RLUs). Sluc, RLUs of firefly luciferase activity in miR-22–transfected sample. Srenilla, RLUs of renilla luciferase activity in miR-22–transfected sample. Cluc, RLUs of firefly luciferase activity in cont miR–transfected groups. Crenilla, RLUs of renilla luciferase activity in cont miR–transfected groups. PC, positive control. (F) Representative Western blot analysis of SIRT1, Sp1, and CDK6 in SiHa and MDA-D3 cells transfected with cont miR (C) and mature miR-22 (22) at 72 h after transfection. (G) The expression levels of SIRT1, CDK6, and pRB phosphorylation in MRC-5 and HMEC184hTERT cells were analyzed by immunoblotting. β-actin was used as a loading control and the relative density of bands was densitometrically quantified. Y, young; S, senescent; hTERT, 184hTERT. Data in all the panels represent mean ± SEM (n = 3). *, P < 0.05; **, P < 0.01.