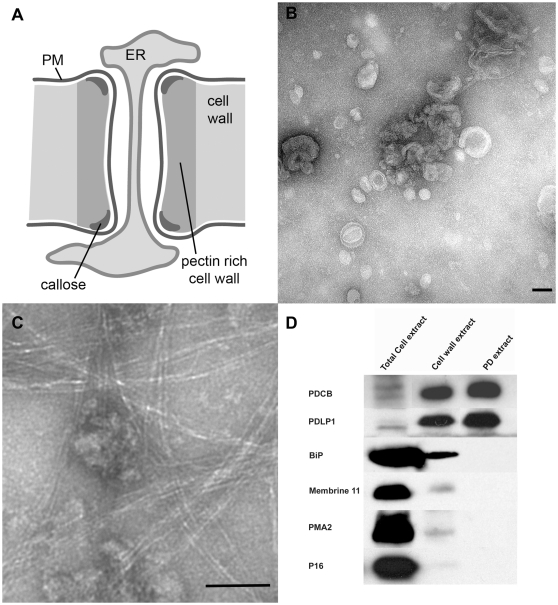
Figure 1. Isolation of plasmodesmata.
The basic structure of plasmodesma (PD) is illustrated in Panel A. In addition to the key physical elements of PM, ER, desmotubule in the wall, a speculative arrangement of actin spiralled around the desmotubule is shown. Panel B shows a negatively stained electron micrograph of membranous PD (pellet P2 in M&M) collected after release from the cell wall following cellulase digestion, while Panel C shows contamination of the PD with residual cell wall fibres, observed very occasionally. Scale bars = 100 nm. Panel D – Immunoblot analysis of fractions harvested during PD isolation procedure. Proteins extracted from whole cells, cell walls (pellet P1 in M&M) and purified PD (pellet P2 in M&M) were analysed using antibodies to the PD marker PDLP1, BiP (ER), Membrine11 (Golgi), PMA2 (PM) and P16 (chloroplast thylakoid envelope). While PDLP was enriched through the isolation procedure, the other proteins diminished and were virtually undetectable in the final PD preparation. Total cell extract: proteins extracted from 6 µl of Arabidopsis cell suspension lysate (corresponding to 0.6 µl of purified cell wall). Cell wall extract: proteins extracted from 75 µl of purified cell walls (pellet P1). PD extract: proteins extracted from 375 µl of purified PD (pellet P2).