Abstract
The Guinea-Bissau family of strains is a unique group of the Mycobacterium tuberculosis complex that, although genotypically closely related, phenotypically demonstrates considerable heterogeneity. We have investigated 414 M. tuberculosis complex strains collected in Guinea-Bissau between 1989 and 2008 in order to further characterize the Guinea-Bissau family of strains. To determine the strain lineages present in the study sample, binary outcomes of spoligotyping were compared with spoligotypes existing in the international database SITVIT2. The major circulating M. tuberculosis clades ranked in the following order: AFRI (n = 195, 47.10%), Latin-American-Mediterranean (LAM) (n = 75, 18.12%), ill-defined T clade (n = 53, 12.8%), Haarlem (n = 37, 8.85%), East-African-Indian (EAI) (n = 25, 6.04%), Unknown (n = 12, 2.87%), Beijing (n = 7, 1.68%), X clade (n = 4, 0.96%), Manu (n = 4, 0.97%), CAS (n = 2, 0.48%). Two strains of the LAM clade isolated in 2007 belonged to the Cameroon family (SIT61). All AFRI isolates except one belonged to the Guinea-Bissau family, i.e. they have an AFRI_1 spoligotype pattern, they have a distinct RFLP pattern with low numbers of IS6110 insertions, and they lack the regions of difference RD7, RD8, RD9 and RD10, RD701 and RD702. This profile classifies the Guinea-Bissau family, irrespective of phenotypic biovar, as part of the M. africanum West African 2 lineage, or the AFRI_1 sublineage according to the spoligtyping nomenclature. Guinea-Bissau family strains display a variation of biochemical traits classically used to differentiate M. tuberculosis from M. bovis. Yet, the differential expression of these biochemical traits was not related to any genes so far investigated (narGHJI and pncA). Guinea-Bissau has the highest prevalence of M. africanum recorded in the African continent, and the Guinea-Bissau family shows a high phylogeographical specificity for Western Africa, with Guinea-Bissau being the epicenter. Trends over time however indicate that this family of strains is waning in most parts of Western Africa, including Guinea-Bissau (p = 0.048).
Introduction
Guinea-Bissau is a West African country with a population of approximately 1.6 million people. It is a country with one of the highest tuberculosis (TB) burdens in the world. In previous studies of Mycobacterium tuberculosis complex isolates from Guinea-Bissau, we identified a unique group of strains that we designated the Guinea-Bissau family of strains [1], [2]. Based on a genotypic analysis of a subset of Guinea-Bissau family isolates, and a comparison of known corresponding genes in mycobacteria outside the M. tuberculosis complex, we proposed that strains of the Guinea-Bissau family belong to a unique branch of the M. tuberculosis complex tree positioned between classical M. tuberculosis and classical M. bovis [2], clustering together within one of the M. africanum subtype I branches [3]. The most intriguing fact about the Guinea-Bissau family of strains is that, although genotypically closely related, they demonstrate a considerable heterogeneity of phenotypic traits [1], [2], with a variation in niacin production, pyrazinamide (PZA) resistance, and nitrate reductase activity, that are basic phenotypic criteria for differentiating M. tuberculosis and M. bovis. Yet, the differential expression of these biochemical traits by the Guinea-Bissau family of strains have not been related to any genes so far investigated [2].
Since the first M. tuberculosis strains from the late 1980s and early 1990s were isolated in Guinea-Bissau [1], [4] there has been a war in the country. This caused the TB program to dissolve, the Reference TB hospital was out of function and the TB laboratory at the Laboratório Nacional de Saúde Pública (LNSP), Bissau, was destroyed and no drug resistance surveillance could be performed. Culture of sputum samples has only recently been made possible again. We have now collected a new sample of M. tuberculosis complex isolates from patients with pulmonary TB and compared them to previously collected (published and unpublished) isolates in terms of genotype. Thus, we hereby reinvestigated M. tuberculosis complex clinical isolates from Guinea-Bissau (n = 414) collected between 1989 and 2008 with emphasis on the phylogeography, of the Guinea-Bissau family of strains, and their differential expression of biochemical traits.
Materials and Methods
Ethics Statement
The Research Committee at Karolinska Institutet exempted this retrospective study from review because it describes bacterial samples collected during the course of routine surveillance, and waived the need for consent due to the fact that the samples received could only be combined with the sex, age, and country of birth of the patients from which the strains were isolated.
Patients
The isolates were collected consecutively as part of epidemiological TB studies in Guinea-Bissau. The patients had been referred to the Raoul Follereau Reference TB hospital in Bissau, Guinea-Bissau from various part of the country. The Hospital is the TB National Reference Center, therefore receives patients from the entire country, namely patients referred from regional hospitals or health centers and also patients coming by themselves. Patients come from the entire country (Bissau, Gabu, Bafata, Cacheu regions).
Study A
During 1989–1994, 229 isolates from the same number of patients with positive cultures for the M. tuberculosis complex were collected as part of an epidemiological TB study in Guinea-Bissau [1], [2], [4], [5], [6].
Study B
As part of a survey of drug resistance, a systematic collection of M. tuberculosis complex isolates was performed between 1994–1998. Approximately 700 sputum samples from the same number of patients with suspected pulmonary TB were collected at the Raoul Follereau Reference TB hospital. Positive culture for the M. tuberculosis complex was confirmed for 192 samples.
Study C
During 2006, 2007 and 2008 approximately 400 sputum samples from the same number of patients were collected. Out of these, 85 were culture positive for the M. tuberculosis complex.
Mycobacterium tuberculosis complex isolates
Isolates included in study A, B and late part of study C were cultured at the LNSP as previously described [1]. Culture in the early part of study C was performed at the TB laboratory, Karolinska University Hospital, Stockholm, Sweden. All M. tuberculosis complex isolates were transported to the Reference TB laboratory at the Swedish Institute for Infectious Disease Control, Solna, Sweden, for further molecular characterization.
Spoligotyping
Spoligotyping relied on the amplification of the polymorphic direct repeat region [7] to obtain hybridization patterns of the amplified DNA using multiple synthetic spacer oligonucleotides, which are covalently bound to a membrane (Isogen Bioscience BV, The Netherlands).
Database comparison of spoligotypes
Spoligotypes in binary format were entered in the SITVIT2 database (Pasteur Institute of Guadeloupe), which is an updated version of the previously released SpolDB4 database [8]. Of 414 isolates 229 had been previously entered by us into the database, and 185 were added in conjunction with this study. At the time of the present study, SITVIT2 contained genotyping information on about 75,000 M. tuberculosis clinical isolates from 160 countries of origin. In this database, SIT (Spoligotype International Type) designates spoligotypes shared by two or more patient isolates, as opposed to “orphan” which designates patterns reported for a single isolate. Major phylogenetic clades were assigned according to signatures provided in SpolDB4, which defined 62 genetic lineages/sub-lineages. These include specific signatures for various M. tuberculosis complex species such as M. bovis, M. microti, M. caprae, M. pinipedii, and M. africanum, as well as rules defining major lineages/sublineages for M. tuberculosis sensu stricto. These include the Beijing clade, the Central Asian (CAS) clade and two sublineages, the East African-Indian (EAI) clade and nine sublineages, the Haarlem (H) clade and three sublineages, the Latin American-Mediterranean (LAM) clade and 12 sublineages, the “Manu” family and three sublineages, the S clade, the IS6110–low-banding X clade and four sublineages, and an ill-defined T clade with five sublineages.
IS6110 RFLP analyses
A standardized method for IS6110 RFLP analysis [9] was used. DNA was extracted and RFLP typing was performed using the insertion sequence IS6110 as a probe and PvuII as the restriction enzyme. Visual bands were analyzed using the BioNumerics software version 5.10 (Applied Maths, Kortrijk, Belgium). Fingerprint patterns were compared by the un-weighted pair-group method of arithmetic averaging using the Jaccard coefficient. Dendrograms were constructed to show the degree of relatedness among strains according to a previously described algorithm [10], and similarity matrixes were generated to visualize the relatedness between the banding patterns of all isolates.
PCR for RD702
Genomic DNA (10–20 ng) for RD702, which delineates the branch of M. africanum West African 2 lineage [11], [12] was amplified in a volume of 25 µL, which contained premixed Taq, Q solution (both from Qiagen), and primers RD702F (TTCCGAGGACCCGTTGTTGAGTGC) and RD702R (GGGCGGGTTGGGTTGCTGGTC) at a final concentration of 0.2 µmol/L [3], [11]. Conditions used for the amplification were as follows: 94°C for 3 min, followed by 35 cycles each of 94°C for 1 min, 64°C for 1 min, and 72°C for 1 min. PCR products of ∼ 2 kb for the control strain H37Rv and ∼ 700 bp for the isolates belonging to M. africanum West African 2 lineage were visualized on a 0.8% agarose gel [12].
Single-nucleotide polymorphism in the narGHJI operon
A mutation in the narGHJI operon has been reported to be responsible for differential nitrate reductase activity of M. tuberculosis (-215T) versus M. bovis (-215C) [13]. The single-nucleotide polymorphism (SNP) at position -215 within the nitrate reductase (narGHJI) operon promoter in M. tuberculosis complex isolates with different NO3 activity was investigated. The isolates were characterized by PCR-RFLP using the previously published primers LC66 (AACCGACGGTGTGGTTGAC) and LC67 (ATCTCGATGGATGGGCGTC) [13]. The PCR products obtained were digested using the restriction endonuclease Sau3AI (Roche-Biomedicals, Meylan, France). This enzyme specifically cuts at the GATC sequence overlapping the M. bovis-like -215C narGHJI promoter site, producing two bands in PCR-RFLP of the LC66-LC67 PCR fragment. In the case of the -215T sequence, the restriction site is absent and as a result the PCR-RFLP of the narGHJI amplicon yields a single band.
Geographical distribution of spoligotypes and lineages
The distribution of spoligotypes in the SITVIT2 database, in relation to their geographical origin was recorded for regions representing ≥5% of a given SIT as compared to their total number in the SITVIT2 database. The various macro-geographical regions and sub-regions were defined according to the specifications of the United Nations (http://unstats.un.org/unsd/methods/m49/m49regin.htm). Correlation of spoligotype families and principal genetic groups (PGG) based on KatG463-gyrA95 polymorphisms [14] was inferred from the reported linking of specific spoligotype patterns to PGG1 (ancient lineages), and PGG2/3 (modern lineages) as previously described [8], [15], [16].
For statistical analysis of trends over time the Chi square test for trend was performed using PASW v 18.
Results
In study A, spoligotyping and RFLP was performed on 229 isolates. In study B, adequate amounts of DNA, in order to perform RFLP, were retrieved from 132 isolates, while it was possible to perform spoligotyping on 138 isolates. In study C, spoligotyping could be performed on 47 isolates, while sufficient amounts of DNA were available for RFLP typing of 41 isolates. In regard to the remaining positive samples they were either too contaminated or the isolates had died during transportation. All isolates that were subjected to RFLP were also spoligotyped. For single-nucleotide polymorphism in the narGHJI operon, 28 isolates were selected from RFLP cluster B:5 [1] in study A.
Spoligotyping
Altogether, 414 isolates were available for spoligotyping, of which 336 isolates were grouped into 42 clusters (2–96 isolates per cluster), accounting for a very high clustering rate of 81.2%, while the remaining 78 (18. 8%) strains did not cluster. Seven clusters included more than ten isolates each.
Study A
Out of 229 isolates, 82 different spoligotypes were obtained, of which 168 (73.4%) were clustered into 21 spoligo-clusters, comprising 2–61 isolates per cluster. The remaining 61 (26.6%) spoligopatterns were unique i.e. the isolates did not cluster with other patient isolates. The 229 isolates are identical to those reported in [1], however a mistake in the recording of one spoligotype (isolate IH-218) has been corrected here. Also, nine isolates with unique spoligotypes (summarized in Table 2 of reference [1]), were not mentioned in the Results section of the 1999 paper.
Study B
Of 138 isolates, 56 different spoligotypes were obtained, of which 98 (71.0%) were clustered into 15 spoligo-clusters, comprising 2–26 isolates per cluster. The remaining 40 (29.0%) spoligopatterns were unique i.e. the isolates did not cluster with other patient isolates.
Study C
Of 47 isolates, 26 different spoligotypes were obtained, of which 29 (61.7%) were clustered into 8 spoligo-clusters, comprising 2–9 isolates per cluster. The remaining 18 (38.3%) spoligopatterns were unique i.e. the isolates did not cluster with other patient isolates.
The spoligotyping results from the SITVIT2 database comparison are summarized in Table 1 . Of 120 different patterns for the 414 strains studied 50 patterns corresponded to orphan strains that were unique among the 75,000 strains recorded in the SITVIT2 database (Supplementary Table S1), as opposed to 70 patterns corresponding to 364 clinical isolates that corresponded to shared-types (Supplementary Table S2), i.e. an identical pattern shared by two or more patients worldwide (within this study, or matching another strain in the SITVIT2 database). A SIT number was attributed to each pattern according to the SITVIT2 database.
Table 1. Sublineage distribution and associated SIT designations for each of the lineages in the study.
| Lineage | Sublineage | SIT |
| AFRI n = 195 (47.10%) | AFRI_1 n = 194 | SIT181 n = 96 |
| SIT187 n = 46 | ||
| orphan n = 26 | ||
| SIT536 n = 6 | ||
| SIT188 n = 3 | ||
| SIT318 n = 2 | ||
| SIT326 n = 2 | ||
| SIT525 n = 2 | ||
| SIT530 n = 2 | ||
| SIT532 n = 2 | ||
| SIT3104 n = 2 | ||
| SIT3118* n = 2 | ||
| SIT324 n = 1 | ||
| SIT537 n = 1 | ||
| SIT3130 n = 1 | ||
| AFRI_3 n = 1 | orphan n = 1 | |
| LAM n = 75 (18.12%) | LAM9 n = 43 | SIT42 n = 36 |
| SIT2201 n = 2 | ||
| SIT3020* n = 2 | ||
| SIT766 n = 1 | ||
| SIT866 n = 1 | ||
| orphan n = 1 | ||
| LAM1 n = 14 | SIT20 n = 11 | |
| SIT534 n = 3 | ||
| LAM4 n = 9 | SIT60 n = 7 | |
| SIT828 n = 1 | ||
| orphan n = 1 | ||
| LAM5 n = 3 | SIT93 n = 1 | |
| orphan n = 2 | ||
| LAM2 n = 2 | SIT604 n = 2 | |
| LAM3 n = 2 | SIT4 n = 1 | |
| SIT3101* n = 1 | ||
| LAM10-CAM n = 2 | SIT61 n = 2 | |
| T clade n = 53 (12.80%) | T1 n = 41 | SIT53 n = 10 |
| SIT244 n = 7 | ||
| SIT334 n = 3 | ||
| SIT804 n = 3 | ||
| orphan n = 4 | ||
| SIT3100* n = 3 | ||
| SIT196 n = 2 | ||
| SIT522 n = 2 | ||
| SIT535 n = 2 | ||
| SIT230 n = 1 | ||
| SIT521 n = 1 | ||
| SIT611 n = 1 | ||
| SIT801 n = 1 | ||
| SIT888 n = 1 | ||
| T n = 8 | SIT73 n = 5 | |
| SIT3129* n = 2 | ||
| SIT2440 n = 1 | ||
| T5 n = 3 | orphan n = 2 | |
| SIT44 n = 1 | ||
| T2 n = 1 | orphan n = 1 | |
| Haarlem n = 37 (8.94%) | H1 n = 22 | SIT47 n = 18 |
| SIT62 n = 1 | ||
| SIT531 n = 1 | ||
| SIT2030* n = 1 | ||
| orphan n = 1 | ||
| H3 n = 15 | SIT50 n = 12 | |
| SIT533 n = 2 | ||
| SIT75 n = 1 | ||
| EAI n = 25 (6.04%) | EAI5 n = 20 | SIT527 n = 9 |
| SIT528 n = 4 | ||
| orphan n = 4 | ||
| SIT458 n = 2 | ||
| SIT236 n = 1 | ||
| EAI-SOM n = 4 | SIT529 n = 3 | |
| SIT538 n = 1 | ||
| EAI6-BGD n = 1 | SIT129 n = 1 | |
| Unknown signatures n = 13 (3.14%) | Unk n = 13 | orphan n = 6 |
| SIT2445 n = 2 | ||
| SIT3131* n = 2 | ||
| SIT523 n = 1 | ||
| SIT1200 n = 1 | ||
| SIT1204 n = 1 | ||
| Beijing n = 7 (1.69%) | Beijing n = 7 | SIT1 n = 7 |
| × clade n = 4 (0.97%) | ×3 n = 2 | SIT92 n = 2 |
| ×1 n = 2 | SIT119 n = 2 | |
| Manu n = 4 (0.97%) | Manu2 n = 2 | SIT54 n = 1 |
| orphan n = 1 | ||
| Manu1 n = 1 | SIT3132* n = 1 | |
| Manu-ancestor n = 1 | SIT523 n = 1 | |
| CAS n = 2 (0.48%) | CAS1-DEL n = 2 | SIT3111 n = 2 |
Newly-defined SITs are shown by an asterisk while the prototypes defining a sublineage in SITVIT2 database are shown in bold italics.
When the overall repartition of strains according to major M. tuberculosis genotypic families was defined ( Table 1 ), the major circulating M. tuberculosis clades ranked in the following order: AFRI (n = 195, 47.10%) > LAM (n = 75, 18.12%) > ill-defined T clade (n = 53, 12.80%) > Haarlem (n = 37, 8.85%) > EAI (n = 25, 6.04%) > Unknown (n = 12, 2.87%) > Beijing (n = 7, 1.68%) > X clade (n = 4, 0.96%) > Manu (n = 4, 0.97%), CAS (n = 2, 0.48%). Two strains of the LAM clade, both isolated in 2007, belonged to the Cameroon family (SIT61) [17].
The most predominant lineage in Guinea-Bissau was AFRI, which accounted for as high as 47.10% of all TB cases. Surprisingly all the cases with AFRI with the exception of a single orphan isolate classified as AFRI_3, were caused by the AFRI_1 sublineage split into two spoligopatterns – SIT181 (the prototype of the AFRI_1 sublineage) and SIT187. As compared to the SITVIT2, these two SITs were highly predominant in Guinea-Bissau since they alone accounted for 36.92% and 52.87% of all such strains in the database (414 of 423 isolates from Guinea-Bissau in the database originating from this study) with extremely high phylogeographical specificity for Western Africa (74.62 and 74.71% respectively, (Supplementary Table S3).
Another interesting observation was a pattern belonging to the EAI5 sublineage (SIT527, n = 9 strains in this study), with 100% of the cases being identified so far exclusively in Guinea-Bissau. These lineages are characteristic of the evolutionary ancient PGG1 strains. The remaining predominant SITs (SIT42 and SIT20 belonging to the LAM9 and LAM1 sublineages of LAM clade, and SIT47 and SIT50 belonging to the H1 and H3 sublineages of Haarlem clade) were characteristic of the evolutionary-recent PGG2/3 subgroups. The last predominant spoligotype (SIT53) also belonged to the evolutionary-recent T1 sublineage, nonetheless, the “T” family does not represent a genotypic lineage in a strict evolutionary sense since it was defined by default to include strains that could not be classified in one of the established lineages with well-established phylogeographical specificity such as the Haarlem, LAM, CAS, and EAI lineages [8].
Thus if one adds all the PGG1 isolates in Guinea-Bissau, i.e., AFRI n = 195, EAI n = 25, Beijing n = 7, Manu n = 4, CAS n = 2, and one unknown lineage pattern (SIT1200) that could be nonetheless classified as PGG1, the total number of ancestral strains in Guinea-Bissau amounts to 234/414 or 56.52% of all strains.
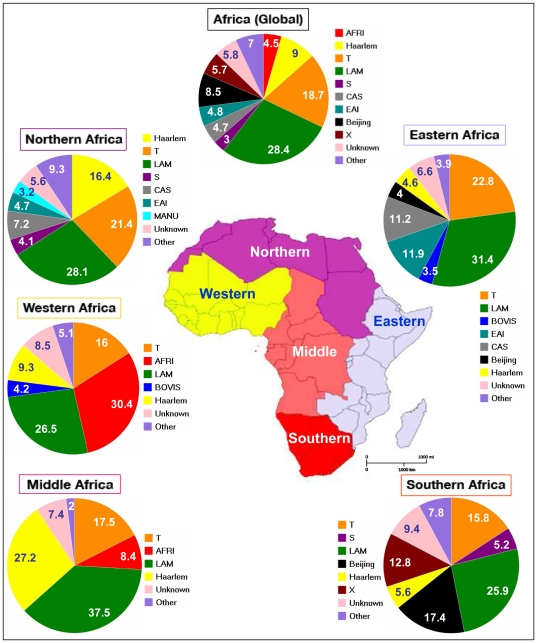
To get an indication of the distribution of the Guinea-Bissau family in Africa, and in spite of the fact that the SITVIT2 is not absolutely representative in terms of proportional representation, we decided to compare all the data obtained in Guinea-Bissau, and compared it to isolates in the SITVIT2 from various subregions of Africa following United Nations geoscheme, i.e., Eastern Africa, Middle Africa, Northern Africa, Southern Africa, and Western Africa (which included Guinea-Bissau). The results obtained for a global collection of 11956 clinical isolates in the database are summarized in Figure 1 , and highlight major differences in the population structure of circulating tubercle bacilli in Africa. It should be noted that to facilitate comparison, results were only tabulated for all major lineages with a cut-off at 3% (i.e., strains belonging to all minor lineages below the cut-off 3% are pooled together and shown as “Other”). Furthermore, all spoligotyping signatures that are not yet associated to a well-defined genotypic lineage in SITVIT2 were designated as “Unknown”.
Figure 1. Geographical distribution of spoligotyping-based genotypic lineages of Mycobacterium tuberculosis in various subregions of Africa (n = 11956 clinical isolates) in the SITVIT2 database.
As for other subregions, this distribution in Guinea-Bissau does not take into account the minor lineages below the cut-off of 3% that included Beijing, X, Manu and CAS lineages.
RFLP
Out of the 402 isolates investigated, 193 different RFLP patterns were obtained, of which 268 (66.7%) isolates were clustered into 61 RFLP-clusters, comprising 2–45 isolates per cluster. The remaining 132 (32.8%) RFLP patterns were unique i.e. the isolates did not cluster with other patient isolates. Of the RFLP-typed isolates with an M. africanum spoligotype 146/193 (75.6%) isolates, and 121/209 (57.9%) of the non-M. africanum isolates were clustered into 24 and 37 RFLP-clusters respectively. Of those with an M. africanum spoligotype 125/193 had 5 bands or less (median 5 bands) and 38/209 of the non-M. africanum isolates had 5 bands or less (median 9 bands).
RD702
Of the 195 isolates with a M. africanum signature 192 isolates were analyzed for RD702; they all showed a PCR product of ∼ 700 bp indicating that RD702 is absent in the Guinea-Bissau family isolates.
Single-nucleotide polymorphism in the narGHJI operon
Twenty-eight Guinea-Bissau isolates, representing all five biovars [4], were investigated. Six of the isolates were nitrate reductase positive, while 22 were negative. The M. tuberculosis H37Rv reference strain served as a control. All 28 isolates showed a single PCR product with a size of approximately 155 bp. For all but one of the Guinea-Bissau family strains, irrespective of nitrate reductase activity, the subsequent digestion with Sau3AI, specifically cutting at the GATC sequence, produced two bands (∼ 90 and ∼ 70 bp) showing that these strains harbored the -215C promoter SNP. For the M. tuberculosis H37Rv reference strain and one Guinea-Bissau strain (nitrate positive) cleavage was not successful, resulting in a single, uncut band of approximately 155 bp.
Trends over time
The proportion of AFRI_1 significantly decreased over time when the three studies A, B and C were compared (P-value = 0.048). Thus in study A 118 (52%) of 229 isolates were AFRI, in study B 60 (44%) of 138 isolates were AFRI, and in study C 18 (38%) of 47 isolates were AFRI. The AFRI_3 isolate was found in 1996 (study B).
Discussion
The Guinea-Bissau family is a member of the M. africanum West African 2 lineage Traditionally, M. africanum strains were defined according to their phenotypic traits and geographic origin, and were accordingly named West African (subtype I) and East African (subtype II) M. africanum [4], [18]. Based on genetic definitions so called M. africanum subtype II strains are now regarded as phenotypic variants of M. tuberculosis [19], [20], [21]. M. africanum subtype I (West African M. africanum) strains are defined on the basis of particular genotypic characteristics that place them in the M. tuberculosis complex phylogenetic tree [2], [19], [22], [23]. Thus, genomic deletions, or regions of difference (RDs), can effectively distinguish them from M. tuberculosis and M. bovis. Isolates of West African M. africanum lack RD9 and are positive for RD12 [19], [20]. We subsequently refer to M. africanum subtype I as M. africanum. M. africanum strains can be further subdivided into two main lineages, West African 1 and West African 2 [12], [20]. West African 1 isolates lack RD711, while West African 2 isolates lack RD 7, 8, 10 and 702 [12], [24]. By spoligotyping M. africanum is defined by loss of spacers 8, 9 and 39. Three sublineages are defined based on finer differences: AFRI_1 by loss of spacers 7–9, and 39, AFRI_2 by loss of spacers 8–12, 21–24, and 37–39, and AFRI_3 by loss of spacers 8–12, and 37–39. These subdivisions correspond to LSP-defined classification as follows: AFRI_1 corresponds to West African 2, while AFRI_2 and AFRI_3 collectively constitute the West African 1 lineages.
This study of M. tuberculosis complex isolates from Guinea-Bissau, collected during two decades, shows that TB in Guinea-Bissau is essentially caused by predominant genotypes characteristic of the Western and Middle African region, with the highest prevalence of M. africanum (AFRI_1/West African 2) in the whole African continent. Forty-seven percent of all isolates were of what we call the Guinea-Bissau family, a member of the M. africanum AFRI_1/West African 2 lineage.
Thus, they have an AFRI_1 spoligotype pattern [1] (this study), they have a distinct RFLP pattern with low numbers of IS6110 insertions [1] (this study) and they lack the four regions of difference RD7, RD8, RD9 and RD10 [2], as well as RD701 [3] and RD702 (this study). This profile classifies the Guinea-Bissau family strains, irrespective of phenotypic biovar, as part of the M. africanum West African 2 lineage [24] or the AFRI_1 sublineage according to the spoligotyping nomenclature. This family shows an extremely high phylogeographical specificity for Western Africa, with Guinea-Bissau being the epicenter.
Other lineages. In addition to the AFRI isolates 25 isolates were of the “ancient” EAI lineage, and the remaining strains were “modern” M. tuberculosis strains. These latter strains most likely represent strain importation from countries where such strains predominate. Only a few isolates were of the Beijing lineage, which is relatively rare in Africa, except for parts of South Africa [25].
Historically, Guinea-Bissau is a former colony of Portugal, a country with a high incidence of TB caused by the classical human M. tuberculosis type. Thus, contacts between the populations of Guinea-Bissau and Portugal have probably been more numerous than contacts between the populations of Guinea-Bissau and other African countries. We therefore speculate that these M. tuberculosis strains may have their origin from within Portugal [1]. Indeed, some of the isolates had RFLP patterns that were identical to isolates from Portuguese TB patients [1].
An interesting observation was the exclusive presence of an EAI5 sublineage (SIT527) in Guinea-Bissau. LAM10-CAM strains were found in a very low proportion, with only 2/414 (0.5%) of such isolates, interestingly both strains belonged to LAM10-CAM prototype SIT61. Yet they are predominant in Middle Africa and Western Africa, with phylogeographical specificity of LAM10-CAM for Cameroon and neighboring countries in West Africa [8], [26], and a recent study showed that LAM10-CAM prototype SIT61 was also the most predominant spoligotype in Cotonou, Benin [27].
Another interesting feature was the total absence of M. bovis in our dataset. Similarly, the finding of four Manu strains in Guinea-Bissau is noteworthy. The "Manu" lineage was initially described as a new family from India in 2004 [28], and later similar strains in small numbers were reported in a study from Madagascar [29]. Soon afterwards, it was tentatively subdivided into Manu-1 (deletion of spacer 34), Manu-2 (deletion of spacers 33–34) and Manu-3 (deletion of spacers 34–36) sub-lineages, with the suggestion that it could represent an ancestral clone of PGG1 strains [8]. More recently, Manu lineage strains were reported from Saudi Arabia, [30] Tunisia [31], and Egypt [32]. In the latter study, Manu lineage strains represented as high as 27% of all isolates, with the new addition of SIT523 (all 43 spacers present) designated as “Manu-ancestor” [32]. However, it was recently suggested that some Manu strains may eventually correspond to a “mixed” pattern due to concomitant Beijing and Euro-American lineage strains (the latter comprising H, LAM, X, and T lineages in spoligotyping defined clades), a possibility that may be investigated by RD105 analysis of Manu pattern isolates [33].
Phenotype and genotype
Although the M. africanum-specific deletions each have putative biological consequence, they do not directly account for these mentioned variable biochemical characteristics. However, genomic variations other than the deletion of large sequence polymorphisms (LSPs) may be relevant for the observed phenotypic variability. Small polymorphisms, single nucleotide polymporhisms (SNPs), gene rearrangements, or genomic regions absent from H37Rv but present in M. africanum (duplications, insertions) are overlooked when applying RD analysis. As a result, phenotypic heterogeneity among M. africanum may be due to unidentified genetic polymorphisms.
Classical M. bovis strains, in contrast to M. tuberculosis strains, are naturally resistant to PZA and lack PZase activity. A point mutation in the pncA gene has been thought to be the cause of the defective PZase activity and PZA resistance in classical M. bovis strains [34]. We have investigated isolates of the Guinea-Bissau family (Study A), and found that irrespective of phenotypic PZase activity, they all had the M. tuberculosis genotype in the pncA gene [1] (data not shown). This was true also for isolates lacking PZase activity, indicating that the variation in phenotypic PZase activity is not due to a variation in the pncA gene.
The differential nitrate reductase (NO3) activity of M. tuberculosis versus M. bovis has been reported to be due to a SNP in the narGHJI operon promoter [13]. The Guinea-Bissau strains tested, except for one, all carried the -215C narGHJI promoter genotype, as defined by the two-band pattern observed in PCR-RFLP. This genotype is present in M. bovis and some ancestral M. tuberculosis [35], and has been connected with a reduced nitrate reductase production. Expression of narGHJI is typically induced under anaerobic conditions, anaerobic nitrate reductase activity being expressed in both -215T and -215C genotypes. However, anaerobic nitrite reductase activity is considerably lower in M. bovis, which carries the -215C genotype, compared to M. tuberculosis. Diagnostic nitrate reductase activity is generally measured as the rapid accumulation of nitrite by the bacilli within two hours. M. bovis typically needs a longer time to accumulate detectable amounts of nitrite from nitrate [13]. Thus, the variable expression of nitrate reductase by M. africanum may be due to the narrow time-detection limit of nitrite defined by the test used and not a genotypic difference between individual M. africanum strains.
The differential expression of these biochemical traits is therefore not related to any genes that have been reported to differentiate between M. bovis and M. tuberculosis. Instead, we hypothesize that Guinea-Bissau strains may more readily switch genes on and off during its adaptation to hosts of different species (and hence broaden their host tropism) [4].
Will the Guinea-Bissau family of strains disappear from West Africa?
This analysis shows that there exists a clear gradient in the African continent; M. africanum being predominant in Western and Middle Africa, EAI and CAS in Northern and Eastern Africa, Beijing and X essentially limited to Southern Africa, while LAM, Haarlem, and T are distributed throughout.
During the 15-year period since isolates were first collected, the proportion of isolates of the Guinea-Bissau family have diminished over time. This is in agreement with trends in some other West African countries, Cameroon [17] and Burkina Faso [36], where the proportion of M. africanum isolates have drastically decreased during the last decades. M. africanum appears at various frequencies in West Africa. In Sierra Leone a recent study identified 23/97 (24%) isolates as M. africanum [37], which appears to be similar to that reported from a study in 1992/1993 [18]. In The Gambia, 38% of isolates were M. africanum [38] and in Ghana 29% [39]. In the 1970's, most cases of TB in Cameroon were caused by M. africanum [40], while three decades later only 9% of cases were reported to be caused by M. africanum [17]. This change appears to be due to the recent expansion of the so-called Cameroon family of M. tuberculosis [17], [26]. In Burkina Faso, M. africanum was not detected in a recent survey [36] although M. africanum strains (defined by phenotypic characteristics) were reported in about 20% of cases two decades earlier [36], [41], [42]. Interestingly, during 2007, two strains of the Cameroon family had found their way into our setting.
The reason for the relative decline of M. africanum in West Africa could be a lower transmission capacity or virulence. There is a considerable uncertainty about the virulence of M. africanum. In studies from The Gambia [11], [38], [43], [44] and Ghana [39] this issue has been addressed. No difference in virulence, as assessed by the severity of radiological presentation, was found between the West African 1 and West African 2, and M. africanum was seen to closely resemble M. tuberculosis in pathology [39]. The pathology in patients with smear positive TB caused by M. africanum (West African 2) were found to be as severe as that of M. tuberculosis patients [44]. However, when comparing M. tuberculosis and M. africanum (West African 2) the rate of progression to disease was higher in household contacts of index patients with M. tuberculosis compared to those with M. africanum infection [43]. Interestingly, M. africanum elicited an attenuated T cell response to early secreted antigenic targets in patients with TB and their household contacts [11].
Another reason for the decline of M. africanum in West Africa could be that certain “modern” lineages possess advantages in their ability to disseminate within a community, in relation to more “ancient” lineages such as M. africanum. Of particular interest in this context is the appearance of Beijing isolates, as well as the recent isolation of two strains of the Cameroon family in our study. Beijing strains have recently been shown to have emerged and rapidly expanded in South Africa [25].
In conclusion the Guinea-Bissau family of strains, irrespective of phenotypic biovar, is part of the M. africanum West African 2 lineage, or the AFRI_1sublineage according to the spoligotyping nomenclature. The variation of biochemical traits classically used to differentiate M. tuberculosis from M. bovis is not related to any genes so far investigated (narGHJI and pncA). The Guinea-Bissau family shows an extremely high phylogeographical specificity for Western Africa, with Guinea-Bissau being the epicenter. Trends over time however indicate that this family of strains is waning in most parts of Africa, including Guinea-Bissau.
Supporting Information
Description of the orphan strains (n = 50) and corresponding spoligotyping defined lineages found among a total of 414 M. tuberculosis clinical isolates from studies performed in Guinea Bissau.
(DOC)
Description of 70 shared types containing 364 M. tuberculosis complex clinical isolates from Guinea Bissau (GNB).
(DOC)
Description of predominant SITs (representing 10 or more strains) in Guinea Bissau (GNB), and their worldwide distribution.
(DOC)
Acknowledgments
We thank the staff at the Laboratório Nacional de Saúde Pública, Bissau, the TB National Reference Hospital “Raoul Follereau”, Bissau and the TB laboratory at the Karolinska University Hospital, Stockholm for culturing assistance. We also thank Biniyam Ghebru for running the narGHJI PCR, Lina Benson for statistical help and Thierry Zozio and Thomas Burguière (Institut Pasteur de Guadeloupe) for helping with mapping of the spoligotype patterns using the SITVIT2 database.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was funded by grants from the Swedish International Development Cooperation Agency, the Swedish Heart-Lung Foundation, and the Swedish Research Council. The SITVIT2 database project benefited from research grants from the Regional Council of Guadeloupe (grant CR/08-1612), and by United States National Institutes of Health (grant R01LM009731). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kallenius G, Koivula T, Ghebremichael S, Hoffner SE, Norberg R, et al. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J Clin Microbiol. 1999;37:3872–3878. doi: 10.1128/jcm.37.12.3872-3878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koivula T, Ekman M, Leitner T, Lofdahl S, Ghebremicahel S, et al. Genetic characterization of the Guinea-Bissau family of Mycobacterium tuberculosis complex strains. Microbes Infect. 2004;6:272–278. doi: 10.1016/j.micinf.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Mostowy S, Onipede A, Gagneux S, Niemann S, Kremer K, et al. Genomic analysis distinguishes Mycobacterium africanum. J Clin Microbiol. 2004;42:3594–3599. doi: 10.1128/JCM.42.8.3594-3599.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffner SE, Svenson SB, Norberg R, Dias F, Ghebremichael S, et al. Biochemical heterogeneity of Mycobacterium tuberculosis complex isolates in Guinea-Bissau. J Clin Microbiol. 1993;31:2215–2217. doi: 10.1128/jcm.31.8.2215-2217.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dias F, Michael SG, Hoffner SE, Martins L, Norberg R, et al. Drug susceptibility in Mycobacterium tuberculosis of a sample of patients in Guinea Bissau. Tuber Lung Dis. 1993;74:129–130. doi: 10.1016/0962-8479(93)90040-5. [DOI] [PubMed] [Google Scholar]
- 6.Naucler A, Winqvist N, Dias F, Koivula T, Lacerda L, et al. Pulmonary tuberculosis in Guinea-Bissau: clinical and bacteriological findings, human immunodeficiency virus status and short term survival of hospitalized patients. Tuber Lung Dis. 1996;77:226–232. doi: 10.1016/s0962-8479(96)90005-2. [DOI] [PubMed] [Google Scholar]
- 7.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Jong BC, Hill PC, Brookes RH, Gagneux S, Jeffries DJ, et al. Mycobacterium africanum elicits an attenuated T cell response to early secreted antigenic target, 6 kDa, in patients with tuberculosis and their household contacts. J Infect Dis. 2006;193:1279–1286. doi: 10.1086/502977. [DOI] [PubMed] [Google Scholar]
- 12.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stermann M, Bohrssen A, Diephaus C, Maass S, Bange FC. Polymorphic nucleotide within the promoter of nitrate reductase (NarGHJI) is specific for Mycobacterium tuberculosis. J Clin Microbiol. 2003;41:3252–3259. doi: 10.1128/JCM.41.7.3252-3259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez MC, Ahmed N, Willery E, Narayanan S, Hasnain SE, et al. Predominance of ancestral lineages of Mycobacterium tuberculosis in India. Emerg Infect Dis. 2006;12:1367–1374. doi: 10.3201/eid1209.050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina-Torres CA, Moreno-Torres E, Ocampo-Candiani J, Rendon A, Blackwood K, et al. Mycobacterium tuberculosis spoligotypes in Monterrey, Mexico. J Clin Microbiol. 48:448–455. doi: 10.1128/JCM.01894-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niobe-Eyangoh SN, Kuaban C, Sorlin P, Cunin P, Thonnon J, et al. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J Clin Microbiol. 2003;41:2547–2553. doi: 10.1128/JCM.41.6.2547-2553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas WH, Bretzel G, Amthor B, Schilke K, Krommes G, et al. Comparison of DNA fingerprint patterns of isolates of Mycobacterium africanum from east and west Africa. J Clin Microbiol. 1997;35:663–666. doi: 10.1128/jcm.35.3.663-666.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemann S, Kubica T, Bange FC, Adjei O, Browne EN, et al. The species Mycobacterium africanum in the light of new molecular markers. J Clin Microbiol. 2004;42:3958–3962. doi: 10.1128/JCM.42.9.3958-3962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asiimwe BB, Koivula T, Kallenius G, Huard RC, Ghebremichael S, et al. Mycobacterium tuberculosis Uganda genotype is the predominant cause of TB in Kampala, Uganda. Int J Tuberc Lung Dis. 2008;12:386–391. [PubMed] [Google Scholar]
- 22.Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J Infect Dis. 2002;186:74–80. doi: 10.1086/341068. [DOI] [PubMed] [Google Scholar]
- 23.Huard RC, Fabre M, de Haas P, Lazzarini LC, van Soolingen D, et al. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J Bacteriol. 2006;188:4271–4287. doi: 10.1128/JB.01783-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–337. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 25.Cowley D, Govender D, February B, Wolfe M, Steyn L, et al. Recent and rapid emergence of W-Beijing strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin Infect Dis. 2008;47:1252–1259. doi: 10.1086/592575. [DOI] [PubMed] [Google Scholar]
- 26.Niobe-Eyangoh SN, Kuaban C, Sorlin P, Thonnon J, Vincent V, et al. Molecular characteristics of strains of the cameroon family, the major group of Mycobacterium tuberculosis in a country with a high prevalence of tuberculosis. J Clin Microbiol. 2004;42:5029–5035. doi: 10.1128/JCM.42.11.5029-5035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Affolabi D, Anyo G, Faihun F, Sanoussi N, Shamputa IC, et al. First molecular epidemiological study of tuberculosis in Benin. Int J Tuberc Lung Dis. 2009;13:317–322. [PubMed] [Google Scholar]
- 28.Singh UB, Suresh N, Bhanu NV, Arora J, Pant H, et al. Predominant tuberculosis spoligotypes, Delhi, India. Emerg Infect Dis. 2004;10:1138–1142. doi: 10.3201/eid1006.030575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferdinand S, Sola C, Chanteau S, Ramarokoto H, Rasolonavalona T, et al. A study of spoligotyping-defined Mycobacterium tuberculosis clades in relation to the origin of peopling and the demographic history in Madagascar. Infect Genet Evol. 2005;5:340–348. doi: 10.1016/j.meegid.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 30.Al-Hajoj SA, Zozio T, Al-Rabiah F, Mohammad V, Al-Nasser M, et al. First insight into the population structure of Mycobacterium tuberculosis in Saudi Arabia. J Clin Microbiol. 2007;45:2467–2473. doi: 10.1128/JCM.02293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Namouchi A, Karboul A, Mhenni B, Khabouchi N, Haltiti R, et al. Genetic profiling of Mycobacterium tuberculosis in Tunisia: predominance and evidence for the establishment of a few genotypes. J Med Microbiol. 2008;57:864–872. doi: 10.1099/jmm.0.47483-0. [DOI] [PubMed] [Google Scholar]
- 32.Helal ZH, Ashour MS, Eissa SA, Abd-Elatef G, Zozio T, et al. Unexpectedly high proportion of ancestral Manu genotype Mycobacterium tuberculosis strains cultured from tuberculosis patients in Egypt. J Clin Microbiol. 2009;47:2794–2801. doi: 10.1128/JCM.00360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viegas SO, Machado A, Groenheit R, Ghebremichael S, Pennhag A, et al. Molecular diversity of Mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in Mozambique. BMC Microbiol. 10:195. doi: 10.1186/1471-2180-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 35.Goh KS, Rastogi N, Berchel M, Huard RC, Sola C. Molecular evolutionary history of tubercle bacilli assessed by study of the polymorphic nucleotide within the nitrate reductase (narGHJI) operon promoter. J Clin Microbiol. 2005;43:4010–4014. doi: 10.1128/JCM.43.8.4010-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Godreuil S, Torrea G, Terru D, Chevenet F, Diagbouga S, et al. First molecular epidemiology study of Mycobacterium tuberculosis in Burkina Faso. J Clin Microbiol. 2007;45:921–927. doi: 10.1128/JCM.01918-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Homolka S, Post E, Oberhauser B, George AG, Westman L, et al. High genetic diversity among Mycobacterium tuberculosis complex strains from Sierra Leone. BMC Microbiol. 2008;8:103. doi: 10.1186/1471-2180-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Jong BC, Hill PC, Brookes RH, Otu JK, Peterson KL, et al. Mycobacterium africanum: a new opportunistic pathogen in HIV infection? Aids. 2005;19:1714–1715. doi: 10.1097/01.aids.0000185991.54595.41. [DOI] [PubMed] [Google Scholar]
- 39.Meyer CG, Scarisbrick G, Niemann S, Browne EN, Chinbuah MA, et al. Pulmonary tuberculosis: virulence of Mycobacterium africanum and relevance in HIV co-infection. Tuberculosis (Edinb) 2008;88:482–489. doi: 10.1016/j.tube.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Huet M, Rist N, Boube G, Potier D. [Bacteriological study of tuberculosis in Cameroon]. Rev Tuberc Pneumol (Paris) 1971;35:413–426. [PubMed] [Google Scholar]
- 41.Ledru S, Cauchoix B, Yameogo M, Zoubga A, Lamande-Chiron J, et al. Impact of short-course therapy on tuberculosis drug resistance in South-West Burkina Faso. Tuber Lung Dis. 1996;77:429–436. doi: 10.1016/s0962-8479(96)90116-1. [DOI] [PubMed] [Google Scholar]
- 42.Rey JL, Villon A, Menard M, Le Mao G, Albert JP. [Present state of resistance to antibiotics by tuberculous bacilli in Africa (author's transl)]. Med Trop (Mars) 1979;39:149–154. [PMC free article] [PubMed] [Google Scholar]
- 43.de Jong BC, Hill PC, Aiken A, Awine T, Antonio M, et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J Infect Dis. 2008;198:1037–1043. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Jong BC, Hill PC, Aiken A, Jeffries DJ, Onipede A, et al. Clinical presentation and outcome of tuberculosis patients infected by M. africanum versus M. tuberculosis. Int J Tuberc Lung Dis. 2007;11:450–456. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of the orphan strains (n = 50) and corresponding spoligotyping defined lineages found among a total of 414 M. tuberculosis clinical isolates from studies performed in Guinea Bissau.
(DOC)
Description of 70 shared types containing 364 M. tuberculosis complex clinical isolates from Guinea Bissau (GNB).
(DOC)
Description of predominant SITs (representing 10 or more strains) in Guinea Bissau (GNB), and their worldwide distribution.
(DOC)