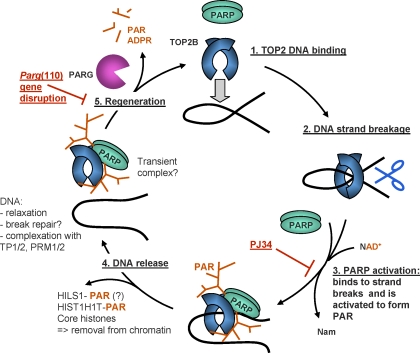
FIG. 6.
Proposed model of TOP2B and PARP1/2 activation coordinating spermatid DNA relaxation with nucleoprotein exchange. TOP2B, and potentially TOP2A, binds to target DNA (1) and generates a DNA strand break (2). Such DNA strand breaks trigger PARP activation and consequently PAR synthesis (3). Modification with PAR leads to catalytic inactivation and release of the topoisomerase-PARP complex from the DNA, along with PAR acceptor proteins, for example, HILS1, HIST1H1T, and core histones (4). Hypothetically, decatenated DNA is now accessible for binding to other available DNA-binding proteins, for example, transition proteins (TP1/2) and protamines (PRM1/2). The PARsylated topoisomerase-PARP complex is released from the DNA, and degradation of PAR by PARG is essential to restore enzyme activities (5). After removing histones and relieving DNA supercoiling, regenerated PARP and TOP2B enzymes can then move to another site along the chromatin and perform the same actions (1). Pharmacological PARP inhibition with PJ34 inhibits PARP activation (red block at step 3), while PAR accumulation observed in the Parg gene-disrupted mouse model indicates partially compromised enzyme regeneration (red block at step 5). ADPR, adenosine diphosphate ribose; Nam, nicotinamide.