Figure 5.
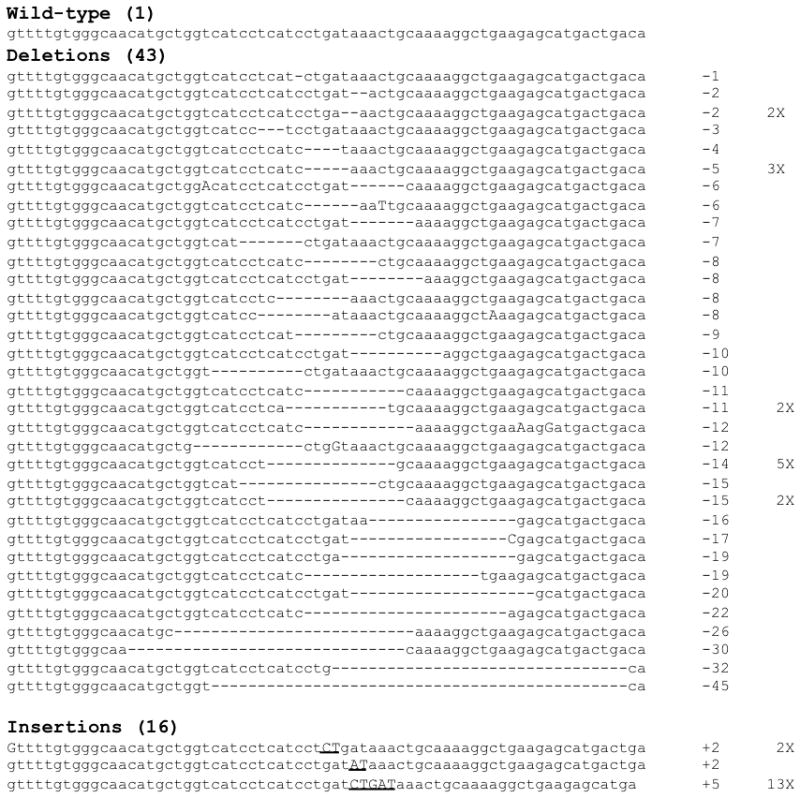
ZFN activity produces heterogeneous mutations in CCR5. Sequence analysis was performed on 60 cloned human CCR5 alleles, PCR amplified from intraepithelial cells from the large intestine of an HIV-infected mouse previously transplanted with ZFN-treated HSC, and at 12 weeks post-infection. The number of nucleotides deleted or inserted at the ZFN target site in each clone is indicated on the right of each sequence, together with the number of times the sequence was found. Dashes (-) indicate deleted bases compared to the wildtype sequence, uppercase letters are point mutations, underlined upper case letters are inserted bases. Some specific mutations of CCR5 occurred more frequently, in particular a 5bp duplication at the ZFN target site that was identified 13 times (bottom sequence). No mutations in CCR5 were observed in a similar analysis performed on control samples from a mouse receiving non-modified HSC (data not shown).
