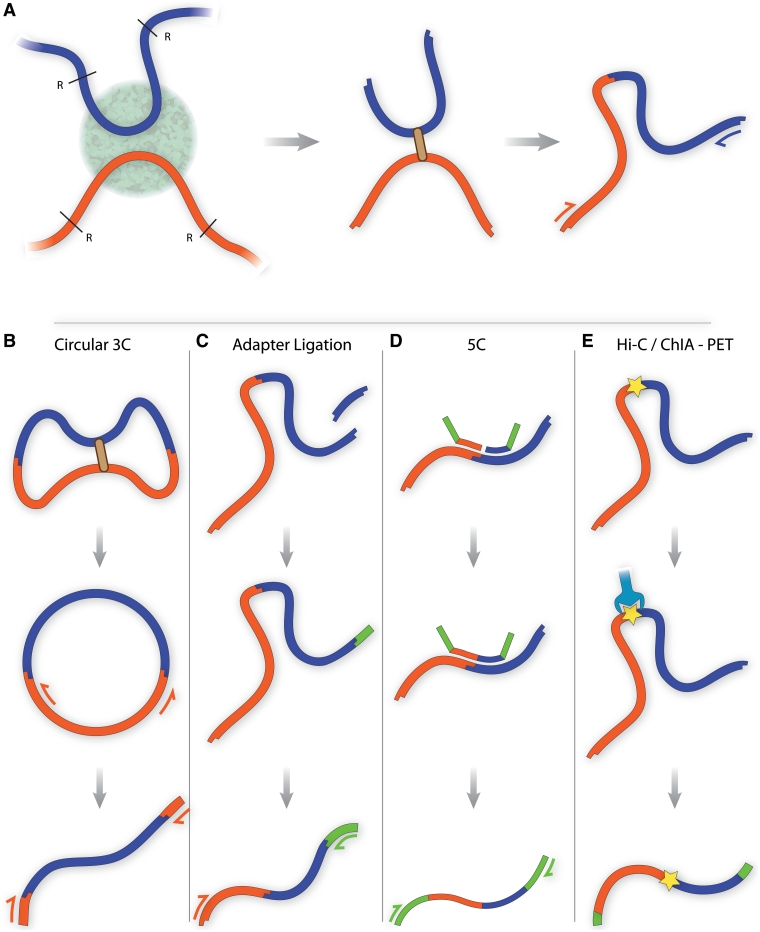
Figure 1:
(A) Traditional chromosome conformation capture (3C). DNA sequences co-localising within a region of the nucleus are fixed using formaldehyde and cut using a restriction endonuclease such as Bgl II or Hind III. Cut DNA ends are joined in dilute conditions, favoring ligation events within a fixed complex. Cross-links are reversed and DNA is purified. PCR and quantitative PCR can be used with specific primers to detect expected products in a quantitative manner. (B) Circularized 3C, as carried out by Zhao et al. and Lomvardas et al. [24, 25]. Würtele et al. and Simonis et al. use a second round of restriction digestion and ligation to reduce the DNA circle size [26, 27] (data not shown). DNA sequences are subjected to a prolonged ligation step to form circles. Nested PCR using primers within the known ‘bait’ sequence is performed, yielding a library with known end fragments containing unknown interacting sequences. (C) Adapter ligation 3C. A second restriction enzyme digestion step creates a sticky end near the ligation junction. An adapter sequence is ligated to this sticky end, enabling PCR amplification of products using primers within the ‘bait’ sequence and adapter. (D) 5C. Multiple ligation-mediated amplification primers containing adapter sequence are annealed directly adjacent to restriction sites of interest. Those paired at ligation junctions are ligated, enabling PCR amplification of the ligation products using the adapter sequences present within the primers. (E) Hi-C/ChIA-PET. All ligation junctions are labeled using a biotin tag. Complexes are purified using streptavidin, and adapters are ligated to the library to enable massive parallel sequencing.