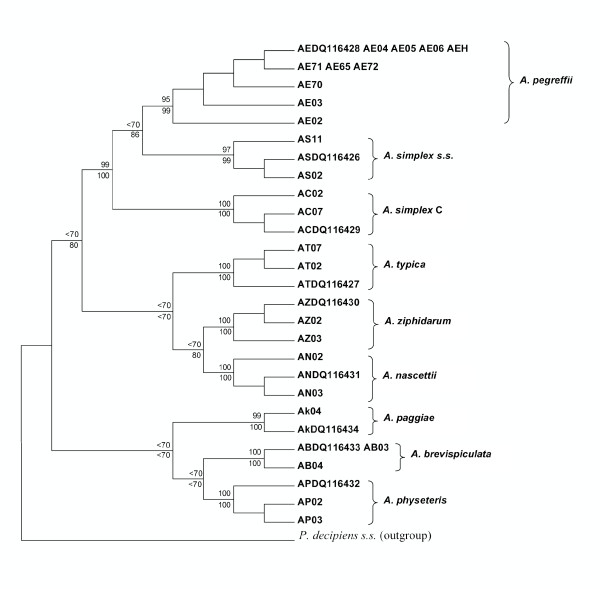
Figure 3.
Cox2 derived Maximum Parsimony (MP) and Neighbour-Joining (NJ) condensed consensus trees inferred by MEGA4.0 for the specimen of Anisakis pegreffii larva from the human intestinal case (code: AEH) sequenced for the mtDNA cox2 (629 bp). The MP tree was obtained by the bootstrap method with a heuristic search. There were 213 polymorphic sites and 178 parsimony-informative sites. Sequences of A. pegreffii from Engraulis encrasicolus are reported with the codes: AE71, AE65, AE72, AE70. Sequences of A. pegreffii from our previous studies are reported with the codes AE03 and AE02 [28]. Number of bootstrap replicates = 1000; bootstrap percentages of the clades ≥ 70 are shown at the nodes, with MP and NJ values are reported above and below the nodes, respectively. Sequences from the other Anisakis spp. are those previously analyzed by us [24,28] and deposited in Genbank. Accession numbers are reported in the text. Pseudoterranova decipiens (s.s.) was used as the outgroup.