Fig. 1.
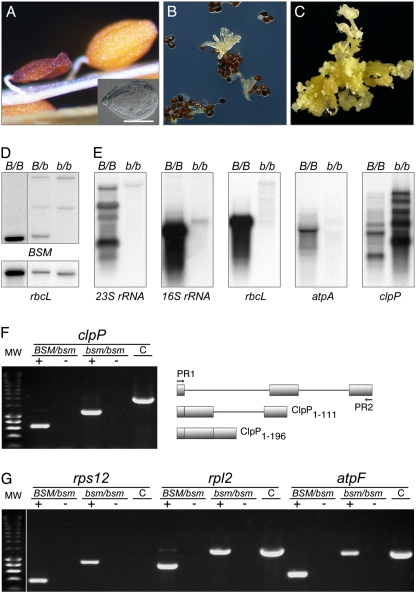
Mutant bsm phenotype and plastid gene expression. (A) Shrunken dry mutant seed, arrested embryo (Inset), and a WT seed. (B) Asynchronous germination of the seeds with arrested embryos. (C) Mixed callus/shoot bsm culture on a medium supplemented with phytohormones. (D) Albino cells are bsm/bsm homozygous. HindIII-digested genomic DNA from albino cells (b/b) was compared with DNA of green homo- (B/B) and heterozygous (B/b) plants by hybridization analysis with radioactive probes detecting nuclear BSM and plastid rbcL genes. (E) Analysis of plastid transcripts. RNA was extracted from green plants (B/B) or bsm albino shoot cultures (b/b). Analyzed plastid genes were rrn23S and rrn16S for ribosomal RNAs (23S rRNA and 16S rRNA), the large subunit of Rubisco (rbcL), the α-subunit of the plastidial ATPase (atpA), and the ClpP subunit of ClpPR protease (clpP). (F) bsm cells do not splice the second clpP group IIa intron. DNA products were amplified by PCR with specific primers (PR1 and PR2) for clpP/AtCg00670. The templates were first-strand cDNA (+), RNA (to control for DNA contamination; −), and genomic DNA (to provide a size marker for a PCR fragment representing fully nonspliced clpP transcript; C). The drawing illustrates the exon–intron gene structure, organization of processed transcripts, and lengths of encoded polypeptides. (G) atpF, rpl2, and rps12 intron splicing. The labeling is as in F. The first group IIb intron of rps12 is encoded by two different genes, which require a transsplicing event for processing, the reason why the PCR product is generated only with cDNA and not with DNA as a template.