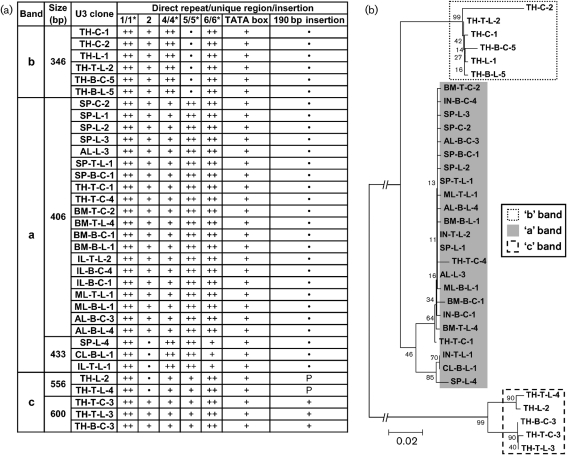
Fig. 2.
Structural features and phylogenetic relationship of the 34 unique U3 sequences isolated from MuLV-ERV genomic RNAs. (a) Sequence features of the 34 unique U3 sequences. Using an alignment protocol, sequence features [direct repeats (1/1*, 4/4*, 5/5* and 6/6*), unique sequences (2 and TATA box) and an insertion (190 bp insertion)] were determined for each U3 sequence (Tomonaga & Coffin, 1998, 1999). a, ‘a’ Band; b, ‘b’ band; c, ‘c’ band; ++, presence of direct repeat; +, presence of single unit; P, partial; •, absence. (b) Phylogenetic relationship of the 34 unique U3 sequences. The phylogenetic tree was established based on the multiple alignment data. The node values indicate the percentage probability for a specific branching. Bar, 0.02 nucleotide substitutions per site.