Abstract
Hypertrophy occurs in urinary bladder wall smooth muscle (BSM) in men with partial bladder outlet obstruction (PBOO) caused by benign prostatic hyperplasia (BPH) and in animal models of PBOO. Hypertrophied BSM from the rabbit model exhibits down-regulation of caveolin-1, a structural and functional protein of caveolae that function as signaling platforms to mediate interaction between receptor proteins and adaptor and effector molecules to regulate signal generation, amplification, and diversification. Caveolin-1 expression is diminished in PBOO-induced BSM hypertrophy in mice and in men with BPH. The proximal promoter of the human and mouse caveolin-1 (CAV1) gene was characterized, and it was observed that the transcription factor GATA-6 binds this promoter, causing reduced expression of caveolin-1. Furthermore, caveolin-1 expression levels inversely correlate with the abundance of GATA-6 in BSM hypertrophy in mice and human beings. Silencing of GATA6 gene expression up-regulates caveolin-1 expression, whereas overexpression of GATA-6 protein sustains the transcriptional repression of caveolin-1 in bladder smooth muscle cells. Together, these data suggest that GATA-6 acts as a transcriptional repressor of CAV1 gene expression in PBOO-induced BSM hypertrophy in men and mice. GATA-6-induced transcriptional repression represents a new regulatory mechanism of CAV1 gene expression in pathologic BSM, and may serve as a target for new therapy for BPH-induced bladder dysfunction in aging men.
Benign prostatic hyperplasia (BPH) is common in aging men and is often associated with lower urinary tract symptoms. Partial bladder outlet obstruction (PBOO) has long been considered a key factor in the mechanism through which BPH causes lower urinary tract symptoms. PBOO-induced lower urinary tract symptoms are associated with bladder wall smooth muscle (BSM) remodeling including hypertrophy, thereby altering bladder contractility.1,2 Obstruction-induced BSM remodeling and hypertrophy are compensatory responses to produce the increased force required to empty urine against the obstruction. These compensatory changes help to maintain close to normal bladder function in some cases. However, in some animals and men, these compensatory changes are not sufficient to restore normal bladder function,1–3 eventually leading to severe bladder dysfunction (decompensation). These compensatory changes are associated with altered expression of contractile proteins and various signaling and regulatory proteins such as Rho-activated kinase and caveolins.3–6
Caveolins are members of a family of integral membrane proteins that are the principal components of caveolae, which are 50- to 100-nm flask-shaped invaginations present in a variety of cells including endothelial cells, adipocytes, cardiac myocytes, and smooth muscle cells (SMCs). Caveolae are a morphologically identifiable subset of lipid rafts that are involved in numerous cellular processes including vesicular transport, cholesterol homeostasis, and signal transduction.7,8 These organelles are believed to function as signaling platforms in a confined cellular microdomain to mediate interaction between receptor proteins and adaptor and effector molecules to regulate signal generation, amplification, and diversification. Caveolin-1 is the primary constituent of caveolae microdomains localized in the cellular membrane.9 Many signaling molecules such as epidermal growth factors, G proteins, G protein–coupled receptors, Src family protein kinases, Ha-Ras, and nitric oxide synthases, which are localized in the caveolae, interact with caveolins.10,11
Studies of transcriptional regulation of CAV1 gene expression have identified two sterol regulatory element–like sequences that control the response of caveolin to low-density lipoprotein and free cholesterol, and the sterol regulatory element–binding protein-1 inhibits caveolin gene transcription in response to cholesterol efflux.12 CAV1 expression is also regulated directly by FOXO (forkhead box class O) transcription factors, and transcriptional activation of the caveolin-1 promoter by FOXO leads in part to attenuation of epidermal growth factor–induced signaling.13
The GATA family of zinc finger transcription factors has an essential role in developmental processes and in tissue- and cell-specific gene transcription.14,15 The GATA family is so named because of its affinity for the consensus DNA-binding motif 5′-(A/T)GATA(A/G)-3′ in target gene promoters and enhancers.16 These transcription factors contain two distinct zinc finger domains with an adjacent highly conserved basic region, which constitutes the DNA-binding domain.17,18 GATA-6 is involved in regulation of contractile protein expression in vascular SMCs, and, thus is an important contributor to maintenance of the differentiated state.19–21 Likewise, GATA-6 is a transcriptional activator of smooth muscle α-actin in human BSM cells.22 However, microarray analysis has demonstrated that GATA-6 regulates genes associated with synthetic SMC phenotypes, which seem to involve cell-to-cell signaling and cell-matrix interactions.23 GATA-6 also regulates gene expression involved in vascular SMC proliferation in response to mitogenic or mechanical stimulation and in vascular injury.14 The GATA-dependent transcriptional pathway is also essential for efficient induction of the hypertrophic response in cultured cardiomyocytes.24
Although expression of GATA-6 mRNA has been reported in murine BSM,25 the functional role of this factor in the pathophysiology of urinary BSM has not been explored. Earlier reports have indicated a role for caveolae in smooth muscle signaling, and the absence of caveolin-1 in these cells leads to failure of caveolae formation.26 Overexpression of CAV1 in cultured cells induces de novo formation of caveolae.27 In bladder smooth muscle signaling,28 caveolae stimulate transforming growth factor-β1 (TGF-β1) signaling,29 and disruption of caveolae enhances the growth inhibitory activity of TGF-β1 in bladder and urethral SMCs.29 Consistent with these findings, caveolin-1 knockout mice fail to form caveolae, resulting in bladder hypertrophy and impaired bladder smooth muscle contraction.30,31 Caveolin-1 protein expression is also reduced in hypertrophic smooth muscle in obstructed rabbit bladder.6 Thus, it is possible that down-regulation of caveolin-1 expression in smooth muscle contributes to bladder dysfunction.
Given the pivotal role of caveolin-1 in regulating smooth muscle contractile activity, loss of caveolin-1 likely has profound effects on the physiologic functions of cells. Several studies have described changes in caveolin-1 expression under various pathologic conditions32–34; however, the mechanisms responsible for mediating these changes in smooth muscle have not been elucidated. Expression of caveolin-1 in hypertrophic smooth muscle was examined using a mouse model of PBOO produced via partial surgical ligation of the urethra and in BSM cells from men with BPH-induced PBOO. There is evidence that GATA-6 acts as a transcriptional repressor of CAV1 gene expression in BSM. Moreover, obstruction-induced overexpression of GATA-6 in hypertrophic BSM contributes to loss of CAV1 gene expression in the mouse model of PBOO and in men with BPH-induced outlet obstruction, leading to urinary bladder dysfunction.
Materials and Methods
Surgical Induction of PBOO in a Mouse Model
All procedures for creating PBOO were approved by the Institutional Animal Care and Use Committee of Children's Hospital and the University of Pennsylvania. Adult male mice were subjected to partial surgical ligation of the urethra, as previously described.35 Sham-operated animals underwent an identical procedure until the suture was tied down, at which point the suture was removed and the abdomen closed. After the desired times, the mice were anesthetized as before, the bladder was harvested, and the BSM layer was separated from the mucosa and serosa.
Human Bladder Samples
After obtaining institutional review board approval (IRB 803645), frozen bladder tissue samples were obtained from Dr. Gomes of the University of Sao Pãolo, Brazil. The tissue samples were identified only by age and disease. Bladder tissue biopsy specimens were collected from patients aged 62 to 78 years who underwent suprapubic prostatectomy to treat BPH. All patients had severe lower urinary tract symptoms and were preoperatively characterized as having overt bladder outlet obstruction. Bladder outlet obstruction was assessed via multichannel urodynamics using the bladder outlet obstruction index. Patients were characterized as having an obstruction when the bladder outlet obstruction index was >40.36 All definitions conform to the standardized terminology of the International Continence Society.37,38
Control BSM samples were acquired from patients undergoing ureteral reimplantation and from nondiseased bladder tissue from patients with bladder cancer undergoing cystectomy. The reasons for ureteral reimplantation in the control group were distal ureteral stenosis and vesicoureteral reflux in one patient each. No patients in the control group exhibited voiding symptoms, had an American Urological Association symptom score higher than 8, or had clinical symptoms of BPH. Age-matched control individuals were used for comparison with the BPH group.
RNA Extraction and RT-PCR
RNA was extracted from murine and human frozen bladder smooth muscle tissue devoid of mucosa and serosa samples using TRIzol reagent (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's protocol. cDNA synthesis was performed using standard procedures.
Protein Extraction and Immunoblot Analysis
Frozen pieces (50 mg) of human and murine bladder muscle tissue devoid of mucosa and serosa were ground into a fine powder. Protein was extracted and analyzed using Western blot analysis as described previously.6 In brief, total protein from each sample was separated using SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF (polyvinylidene difluoride) membranes (Millipore Corp., Bedford, MA). The membranes were incubated with polyclonal anti-caveolin-1 (Abcam, Inc., Cambridge, MA). Immunoreactive proteins were visualized using an appropriate peroxidase-conjugated secondary antibody using Super Signal Reagent (Pierce Chemical Co., Rockford, IL). Equal loading between lanes was confirmed by probing the membranes with anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody. Nuclear extracts were prepared from mouse and human BSM and from primary BSM cells as described,39 and immunoblot analysis was performed as described previously6 using anti-GATA-5, -GATA-6 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and -p97 antibodies (RDI Research Diagnostics, Inc., Flanders, NJ).
Immunohistochemistry
Frozen sections of BSM tissue from healthy and PBOO mice and from healthy and BPH-induced human BSM tissues were used for preparation of 5-μm sections and mounted on a glass slide. IHC of the bladder sections was performed as described previously.40 Smooth muscle–specific markers such as SM22 and myosin heavy-chain 1 were used to determine the smooth muscle–specific caveolin-1 and GATA-6 in murine and human bladders, respectively. The primary antibodies used were rabbit polyclonal anti–caveolin-1 conjugated with Cy3 (C3990) and mouse monoclonal antibody to smooth muscle myosin heavy-chain I (M7786) (both from Sigma-Aldrich, St. Louis, MO), and goat polyclonal antibody to SM22 (ab10135) and rabbit polyclonal antibody to GATA-6 (ab22600) (both from Abcam, Inc.).
Confocal Microscopy
Laser scanning confocal immunofluorescent microscopy was used to determine the localization of caveolin-1 and GATA-6 in murine and human BSM tissues, respectively. Images were acquired using a Zeiss 2-photon Confocal Zeiss Axiovert 200M inverted microscope (Carl Zeiss Ltd., Welwyn Garden City, Hertfordshire, England). Image series were collected as Z stacks through a sample (human BSM stained with GATA-6 antibody) as a series of time-lapse Z stacks. The rabbit polyclonal antibody to caveolin-1 and anti-mouse IgG secondary antibody were conjugated to Cy3 fluorophore; therefore, their fluorescence emission was collected using the red channel. GATA-6 and SM22 were detected using fluorescein isothiocyanate–labeled secondary antibodies; therefore, their fluorescence emission was collected using the green channel. Nuclei were stained using DAPI and assigned to appear as blue.
Promoter Pull-Down Analysis
The promoter pull-down approach was used as described previously41 to identify the proteins that bind to murine CAV1 promoter. To obtain the efficient binding of promoter DNA to the affinity column, the CAV1 promoter (1.1 kb) was divided into smaller regions (400 bp each). PCR-amplified murine CAV1 promoter, +1 to −400, −400 to −800, and −800 to −1100 bp regions were end-labeled using T4 polynucleotide kinase and were coupled to cyanogen bromide–activated Sepharose 4B individually as described previously42 to produce an affinity column. Nuclear proteins from sham and PBOO murine BSM tissue were prepared as described previously,43 and were used for CAV1 DNA affinity column chromatography. Approximately 1 mg nuclear protein was loaded per DNA-Sepharose column, as described previously,41 and the DNA-bound proteins were eluted with elution buffer containing progressive salt gradient (0.25, 0.5, 0.75, and 1 mol/L KCl). The column-eluted fractions were assayed for their ability to bind to their consensus DNA oligonucleotides using a transcription factor enzyme-linked immunosorbent assay (ELISA). The transcription factor ELISA (Active Motif, Inc., Carlsbad, CA) was performed according to the manufacturer's instructions. Proteins present in the eluted fractions were resolved using SDS-PAGE and silver stained using a Silver Stain Plus kit (Bio-Rad Laboratories, Inc., Hercules, CA). Immunoblots of the column purified fractions were developed using Super Signal West Femto maximum sensitivity substrate (Pierce Chemical Co.). The immunoblot with antibody to nuclear p97 represents 1% of the total protein loaded onto the CAV1 promoter DNA affinity column.
ChIP Assays
Chromatin immunoprecipitation (ChIP) assays were performed on bladder smooth muscle tissues from men with or without BPH-induced obstruction and healthy and obstructed mice and on cultured BSM cells according to the procedure originally described by O'Neill and Turner.44 Chromatin samples (200 to 600 bp) were immunoprecipitated using anti–GATA-6 polyclonal antibody (sc-9055) or normal rabbit IgG (Santa Cruz Biotechnology, Inc.) using protein G magnetic beads overnight at 4°C. Beads were collected, and the DNA was eluted as described.44 As an input control, an aliquot of sheared chromatin was purified using a QIAquick PCR purification kit (Qiagen Inc., Valencia, CA). All samples were treated with proteinase K (1 mg/mL) for 1 hour and subjected to PCR using the primer sets given in Table 1. PCR products were subjected to agarose gel electrophoresis and visualized via ethidium bromide staining. DNA band intensities were quantified from the agarose gels, and the data were analyzed via normalizing with the corresponding input values.
Table 1.
Primers Used in the Study
| Gene name | Sequence | Gene accession number |
|---|---|---|
| Murine caveolin-1 mRNA | F: 5′-GCATGTCTGGGGGCAATAC-3′ | AY439333 |
| R: 5′-TTGACCACGTCGTCGTTGA-3′ | ||
| Murine GATA-6 mRNA | F: 5′-ATGCTGTCCGGCCTGCCCTA-3′ | NM_010258 |
| R: 5′-ATGCTGTCCGGCCTGCCCTA-3′ | ||
| Murine caveolin-1 promoter | F: 5′-CAATAAGACAAAGGAGGCTG-3′ | AF124227 |
| GATA-6 binding site | R: 5′-TGTGCTTGACTGTGAGAGAAT-3′ | |
| Human GATA-6 mRNA | F: 5′-CAGTGGCGACTGCGCTGACA-3′ | NM_005257 |
| R: 5′-CTGGCTGGAGTGGCCCAAGC-3′ | ||
| Human caveolin-1 mRNA | F: 5′-CAGCATGTCTGGGGGCAAAT-3′ | NM_001753 |
| R: 5′-TCAGCTCGTCTGCCATGGCC-3′ | ||
| Human caveolin-1 promoter | F: 5′-ACTGCATATGGAAGCATAAG-3′ | AJ133269 |
| GATA-6 binding site | R: 5′-TCAGCTCGTCTGCCATGGCC-3′ | |
| Murine GATA-5 mRNA | F: 5′-GCGCCTCGTACCCCACAACC-3′ | NM_008093 |
| R: 5′-TAGCGGTCGGTTGACCCCGT-3′ | ||
| Human GATA-5 mRNA | F: 5′-AACAGCCTGGAACAGACCACC-3′ | NM_080473 |
| R: 5′-TCCCTCACCAGCCTTCTTGC-3′ | ||
| Murine GAPDH | F: 5′-AGGCCGGTGCTGAGTATGTC-3′ | M32599 |
| R: 5′-TGCCTGCTTCACCACCTTCT-3′ | ||
| Human GAPDH | F: 5′-AGAAGGCTGGGGCTCATTTG-3′ | BC004109 |
| R: 5′-AGGGGCCATCCACAGTCTTC-3′ |
F, forward primer; R, reverse primer.
Primary BSM Cell Preparation
Primary SMCs from mouse bladders were prepared as follows. The bladder urothelial and serosal layers were removed from 8-week-old mice, and the remaining muscle layer was dissected into small pieces. SMCs were dissociated with 0.1% highly purified collagenase (Sigma-Aldrich), and the isolated cells were cultured in M199 supplemented with 10% fetal calf serum, 1%vitamins, and 1% antibiotics-antimycotics. Cultured cells were identified as SMCs by the presence of smooth muscle–specific actin and myosin using immunofluorescence and immunoblot analyses, respectively. Human primary BSM cells were obtained from Lonza Walkersville, Inc., Walkersville, MD.
Plasmid Constructs, Cloning, Transient Transfection, and Promoter-Reporter Gene Assays
Wild-type (WT) murine GATA6 sequence was amplified using PCR, and the adenoviral construct expressing GATA6 was prepared as described previously.45 In addition, the PCR-amplified murine GATA6 cDNA was cloned into the pCMV4 vector. GATA-5 cDNA (OriGene Technologies, Inc., Rockville, MD) was used in cotransfection experiments with CAV1 promoter. Mutation in the GATA-6 binding site of the murine and human CAV1 promoters was introduced using a site-directed mutagenesis kit (Quick-Change II XL; Stratagene Corp., La Jolla, CA).
Plasmids were transfected into mouse and human bladder primary SMCs via electroporation using Nucleofactor II (Amaxa, Lonza Walkersville, Inc., Walkersville, MD) according to the manufacturer's instructions. In all transfections, Renilla luciferase was included to normalize for transfection efficiency. Firefly and Renilla luciferase activities were determined using a reporter assay system (Dual-Luciferase; Promega Corp., Madison, WI). Cleared extracts (100 μL per well) were prepared using the passive lysis buffer for dual luciferase assays, and luciferase activity was measured according to the manufacturer's instructions.
Adenovirus Transduction, RT-PCR, and Immunoblot Analysis
Mouse and human BSM cells were seeded into six-well plates at a density of 2 × 105 cells per well and grown overnight to near confluence. Cells were washed with PBS and infected with adenovirus encoding LacZ or GATA-6 at a multiplicity of infection of 100. Cells were harvested 72 hours after infection, and Western blot analysis was performed as described previously.6
GATA6 Gene Silencing by RNA Interference
Endogenous gene expression of GATA6 was silenced by transient transfection of predesigned small interfering RNA (siRNA) lentivirus particles for mouse GATA-6 (sc-37908V), human GATA-6 (sc-37907V), and scrambled siRNA (sc-108080) (all from Santa Cruz Biotechnology, Inc.). The transfection experiments were performed according to the manufacturer's instructions. After 72 hours, total cell lysates were analyzed for knockdown of GATA-6 mRNA and protein levels using RT-PCR and immunoblot analysis, respectively. A parallel set of transfected cells were used for ChIP assays. In co-transfection experiments, the CAV1 promoter reporter plasmid was introduced into SMCs along with the GATA-6 siRNA. Murine and human bladder primary SMCs (1 × 106) were transiently transfected with double-stranded murine GATA-6 (sc-37908), human GATA-6 (sc-37907), and scrambled siRNA (sc- 37007) at a final concentration of 25 nmol/L via electroporation using Nucleofactor II (Amaxa). The endogenous GATA5 gene expression was silenced with GATA-5-short-hairpin RNA (shRNA) plasmid (sc-35456-SH) via electroporation in human BSM cells
Statistical Analysis
Where appropriate, comparisons between experimental groups were performed using Student's t-test. P < 0.05 was considered statistically significant.
Results
Obstruction-Induced Bladder Hypertrophy Down-Regulates Caveolin-1 mRNA and Protein Expression in Smooth Muscle
Earlier work from our laboratory has demonstrated that caveolin-1 protein expression is down-regulated in bladder smooth muscle from obstruction-induced hypertrophy in the rabbit model of PBOO.6 To extend this observation to mice and human beings, caveolin-1 mRNA and protein expression was measured in BSM tissues from PBOO mice over the course of 14 days and from human bladder tissues from patients with BPH-induced PBOO. Compared with controls, mean (SD) caveolin-1 mRNA was decreased 2.8-fold (0.8-fold) by day 14 (Figure 1, A and C; P < 0.01), and caveolin-1 protein content in BSM from PBOO mice decreased 2.5-fold (0.5-fold) during the same period (Figure 1, B and D; P < 0.01). Similarly, caveolin-1 mRNA and protein were down-regulated in BSM from the bladders of patients with BPH compared with healthy human bladders (Figure 2, A–D). These results confirm that caveolin-1 expression is reduced in mice and human beings with PBOO and that repression of CAV1 gene expression is mediated at the transcriptional level in vivo.
Figure 1.
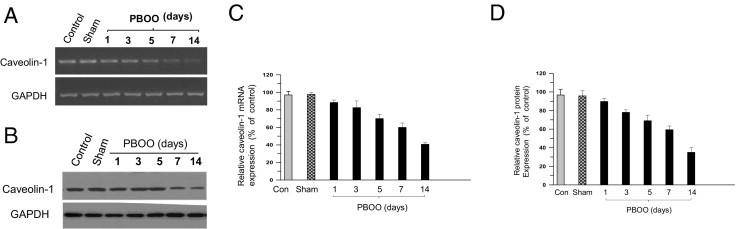
Repression of caveolin-1 (CAV1) gene expression in obstruction-induced hypertrophic BSM in the mouse model of PBOO. A: RNA was extracted from control, sham, and PBOO mouse bladders (n = 10 in each group) up to 14 days after induction, and caveolin-1 was amplified at RT-PCR. B: Protein was extracted from the same samples as in A and was probed at immunoblotting with anti-caveolin-1. C and D: Quantization of band intensity (upper panels in A and B) reported as caveolin-1 expression relative to GAPDH expression (lower panels in A and B) as mean (SD).
Figure 2.
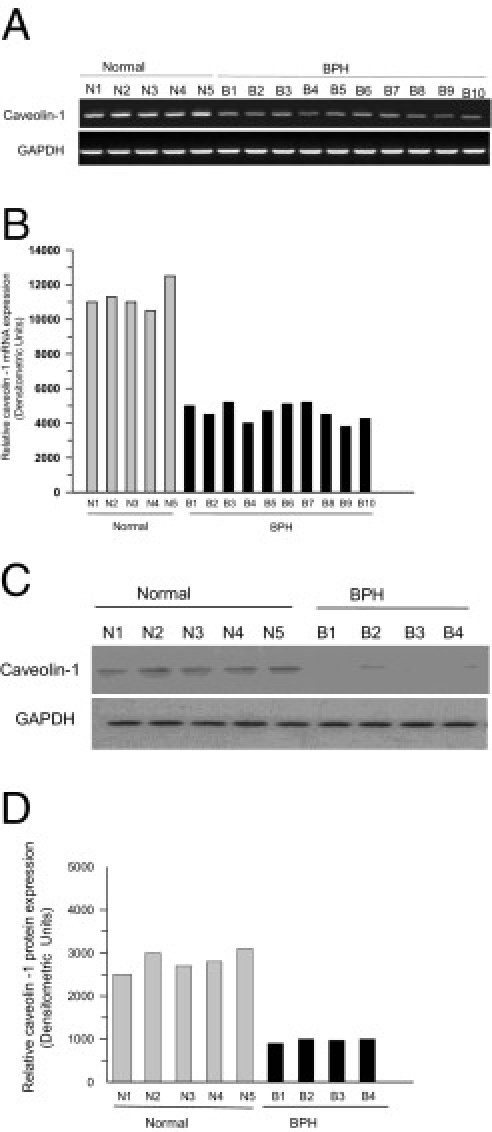
Repression of caveolin-1 (CAV1) gene expression in obstruction-induced hypertrophic BSM in men with BPH-induced PBOO. A: RNA was extracted from healthy and BPH human bladders, and caveolin-1 was amplified at RT-PCR. mRNA and protein expression of caveolin-1 relative to GAPDH is shown for individual bladders. B: Band intensities from A reported as densitometric units of caveolin-1 expression. C: Protein was extracted from the same samples as in A and was probed at immunoblotting with anti-caveolin-1. D: Band intensities from C are given as densitometric units.
In Situ Localization of Caveolin-1 in BSM Cells
Experiments were performed to assess the cellular localization of caveolin-1 protein in bladder samples from mice with PBOO. Bladders were harvested, and tissue sections were stained with anti–caveolin-1 antibody. Representative confocal images obtained from BSM of sham and obstructed mice are shown in Figure 3. Bladder tissues were also stained with smooth muscle–specific protein SM22 to determine the loss of caveolin-1 protein expression in SMCs in obstructed bladders compared with its expression in murine sham bladders. Caveolin-1 primarily localized on the plasma membrane of SMCs in sham bladders (Figure 3, A and C), and its expression was dramatically reduced in SMCs from murine PBOO bladders (Figure 3, B and D). Sections preincubated with caveolin-1 peptide (used for raising the antibody) before staining did not fluoresce (data not shown). These data demonstrate that there is loss of caveolin-1 protein expression in bladder SMCs in PBOO-induced hypertrophy.
Figure 3.
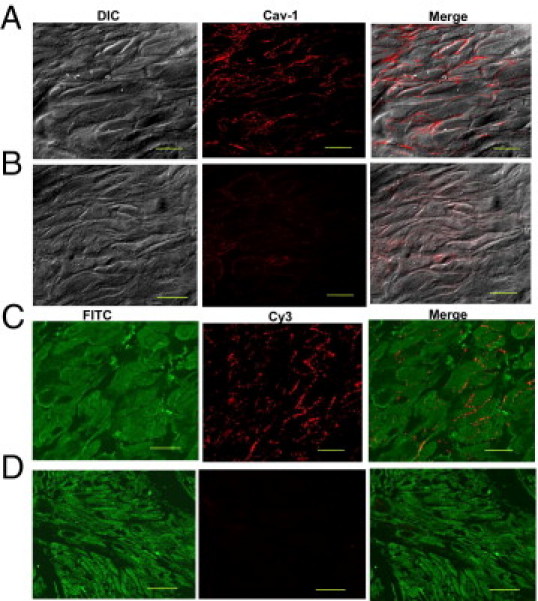
Decreased expression of caveolin-1 protein in obstructed BSM. Bladder sections were prepared from sham (A and C) and PBOO (B and D) mice and were stained with anti–caveolin-1 (Cy3, red) and SM22 (FITC, green). Negative control sections made using antibody preabsorbed with the antigen used for raising the antibody did not reveal immunostaining of the muscle cells (data not shown). Scale bars: 15 μm (A and B); 10 μm (C and D). A and B: Differential interference (DIC), immunofluorescence (Cav-1), and co-localization (Merge) images. FITC-labeled SM22 antibody, Cy3-labeled caveolin-1 antibody, and merged images of smooth muscle bundles from normal (C) and obstructed bladders (D). Sections from obstructed bladders were taken from animals exhibiting various levels of down-regulation of caveolin-1. Cells stained with caveolin-1 also reacted with antibody against smooth muscle–specific protein SM22, indicating that these cells are phenotypically SMCs.
Identification of CAV1 Promoter–Binding Proteins
An attempt to elucidate the molecular basis of CAV1 gene repression in PBOO-induced BSM hypertrophy was performed by searching for transcription factors that bind to the CAV1 promoter. By using bioinformatics tools and programs including Match Public and Transcription Element Search software,46 which use the transcription factor database TRANSFAC, various transcription factor binding sites were identified on both human (Figure 4, A and B) and mouse (Figure 4, C and D) CAV1 promoters, as shown in the schematic representation of the CAV1 gene. This analysis demonstrated that the human CAV1 promoter contains the putative binding sites for the transcription factors GATA-6, ETS, SRE, p53, and SP1, whereas only those for GATA-6, ETS, and SP1 are found in mice.
Figure 4.
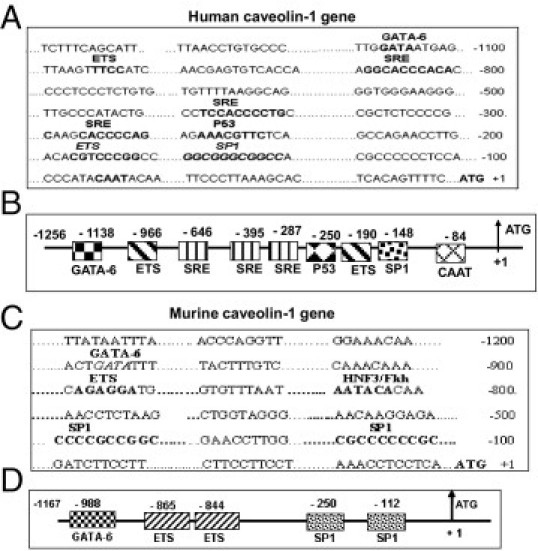
Structural analysis of the caveolin-1 (CAV1) promoter and the schematic representation of the CAV1 gene. The putative binding site for indicated transcription factors on human (A and B) and murine (C and D) CAV1 promoters are shown. Structural details of the human (B) and murine (D) CAV1 gene are indicated. The caveolin-1 construct encompasses nucleotides −1256 to +1 for the human sequence and −1167 to +1 for the mouse sequence, relative to their respective translational start sites.
PBOO Promotes GATA-6 Binding to CAV1 Proximal Promoter GATA-6 Consensus Sites
An attempt was made to identify the transcription factors that are recruited to CAV1 promoter in BSM isolated from sham and PBOO murine bladders. The promoter pull-down approach was used41,42 (Figure 5A) to identify the recruitment of transcription factors to CAV1 promoter and to compare sham and PBOO murine BSM tissue insofar as the binding intensity of the identified transcription factor. The CAV1 promoter regions were conjugated to CNBr-activated Sepharose (see Materials and Methods) to produce the affinity columns. Approximately 1 mg nuclear protein from sham and PBOO mouse BSM tissue samples was loaded on each column, and the bound proteins were eluted with buffer containing salt (0, 0.25, 0.5, 0.75, and 1.0 mol/L KCl). Equal loading to the column was ensured by Western blotting of 1% of each of the nuclear protein samples using p97, which is typically used as a marker for nuclear protein (Figure 5B).
Figure 5.
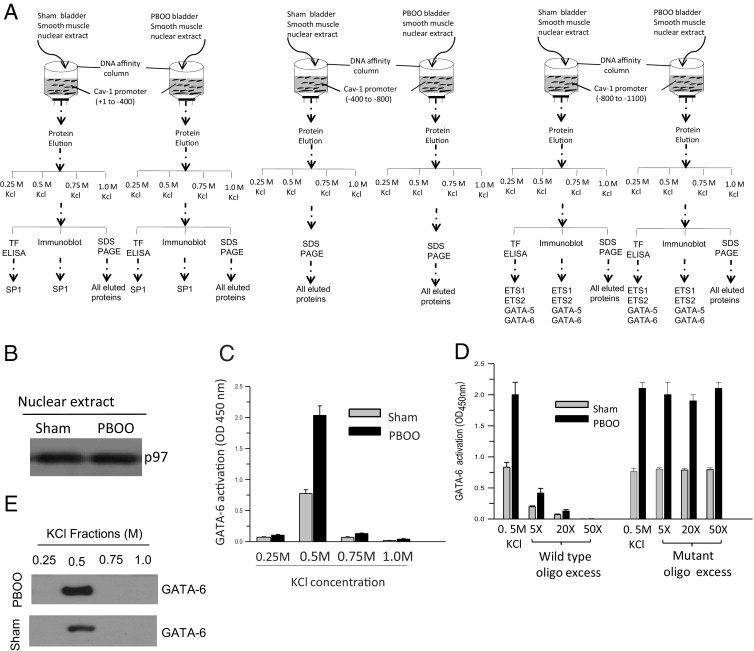
PBOO up-regulates GATA-6 DNA binding activity. A: Schematic representation of caveolin-1 (CAV1) promoter DNA affinity column chromatography using three regions: +1 to −400, −400 to −800, and −800 to −1100. B: Immunoblot analysis of nuclear proteins from sham and PBOO murine bladder with antibody to nuclear p97. C: Protein fractions eluted from the −800 to −1100 bp region of CAV1 promoter DNA affinity column were assayed using a transcription factor ELISA kit, and GATA-6 binding from sham (gray bars) and PBOO (black bars) bladders was expressed as absorbance units at 450 nm. Data are representative of at least three independent experiments, each performed in triplicate. D: Wild-type or mutant GATA-6 binding consensus DNA sequence was added in increasing amounts to the ELISA plate in the oligonucleotide competition assay. The 0.5 mol/L KCl denotes the protein fractions eluted from DNA affinity column using −800 to −1100 bp promoter regions of caveolin-1. E: Immunoblot analysis of protein fractions eluted from the −800 to −1100 bp region of CAV1 promoter DNA affinity column with GATA-6 antibody.
Transcription factors predicted to bind to murine CAV1 promoter were evaluated for their presence in the fractions that were eluted from caveolin-1 DNA affinity column chromatography at immunoblotting, transcription factor ELISA, and SDS-PAGE. Among the ETS family of transcription factors, only Ets-1 and Ets-2 are detected in human and murine bladder smooth muscle nuclear extract. Similarly, among the GATA family transcription factors, GATA-5 and GATA-6 are detected in murine and human bladder smooth muscle nuclear extract. Therefore, the column purified fractions were analyzed for the presence of SP1, Ets-1, Ets-2, GATA-5, and GATA-6 at immunoblot analysis. In parallel, the same fractions were also evaluated for their ability to bind to their consensus DNA oligonucleotide sequence using a transcription factor ELISA. In addition, the column purified fractions were subjected to SDS-PAGE analysis, and the gels were exposed to silver stain to visualize all of the proteins bound to CAV1 promoter.
The fractions eluted with 0.5 mol/L KCl containing elution buffer from the −800 to −1100 bp region of the caveolin-1 DNA affinity column chromatography detected GATA-6 transcription factor as demonstrated using a transcription factor ELISA (Figure 5, C and D) and immunoblot analysis (Figure 5E). An excess of WT (5X, 20X, and 50X) but not mutant (mutation at GATA sequence) consensus oligonucleotides competed with the oligonucleotide sequence immobilized on the plate for binding to GATA-6 in the transcription factor ELISA. Quantitative analysis of the transcription factor ELISA revealed increased binding of GATA-6 to its DNA sequence with the nuclear extract isolated from murine PBOO bladder smooth muscle tissues compared with sham animals. The fractions eluted with 0.25, 0.5, 0.75, and 1.00 mol/L KCl containing elution buffer from the −800 to −1100 bp region of the caveolin-1 DNA affinity column chromatography did not detect GATA-5, Ets-1, and Ets-2 at immunoblot analysis and transcription factor ELISA, and these fractions did not exhibit any detectable protein bands at silver stain SDS-PAGE analysis (data not shown). Similarly, the fractions eluted with 0.25, 0.5, 0.75, and 1.00 mmol/L KCl containing elution buffer from the +1 to −400 bp region of the caveolin-1 DNA affinity column chromatography did not detect SP-1 at immunoblot analysis or transcription factor ELISA, and these fractions failed to demonstrate any detectable protein bands at silver stain SDS-PAGE analysis (data not shown). The fractions eluted with 0.25, 0.5, 0.75, and 1.00 mmol/L KCl containing elution buffer from the −400 to −800 bp region of the caveolin-1 DNA affinity column chromatography did not demonstrate any detectable protein bands at silver stain SDS-PAGE analysis (data not shown).
The enhanced binding of GATA-6 to the GATA motif on the CAV1 proximal promoter in PBOO bladder SMC was confirmed using ChIP assays. In this experiment, chromatin samples were prepared from control, sham, and PBOO bladders, precipitated with anti-GATA-6 antibody, and subjected to PCR amplification of the GATA-6 consensus motif in the promoter. Samples immunoprecipitated with anti-GATA-6 antibody yielded prominent PCR products in PBOO (Figure 6A, lane 9) as compared with sham (Figure 6A, lane 6) and control (Figure 6A, lane 3) mouse bladders. However, samples immunoprecipitated with preimmune rabbit IgG failed to yield PCR products (Figure 6A, lanes 2, 5, and 8). The relative intensities of the bands in Figure 6A are reported as percentage of input chromatin (Figure 6B). Chromatin prepared from BPH human bladder samples also resulted in enhanced PCR amplification of the GATA motif in the human CAV1 proximal promoter after immunoprecipitation with anti–GATA-6 (Figure 6, C and D), confirming that this interaction occurs across species.
Figure 6.
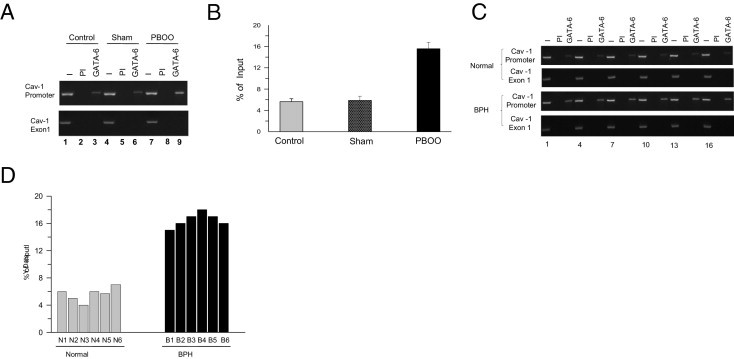
GATA-6 immunoprecipitates with the GATA-6 consensus motif in the caveolin-1 (CAV1) promoter at ChIP analysis. A: Chromatin samples were prepared from control, sham, and PBOO mouse bladders, followed by immunoprecipitation with anti-GATA-6 or preimmune serum. Precipitated fragments were PCR-amplified using primers specific for GATA-6 binding motif on mouse CAV1 promoter (upper panel) or CAV1 exon-1 (lower panel) as negative control. B: Intensities of the bands (shown in A) are presented as percentage of input chromatin (average of six healthy, six sham, and six PBOO samples). C: Chromatin samples were prepared from healthy and BPH human bladder samples as in A. Precipitated fragments were PCR-amplified using primers specific for GATA-6 binding motif on human CAV1 promoter. D: Intensity of the bands (C) is presented as percentage of input chromatin (six healthy men and six men with BPH-induced PBOO, shown individually). I, input; PI, precipitation with preimmune serum.
PBOO Up-Regulates GATA-6 Expression in Mouse PBOO and Human BPH Bladder Samples
The data show that there is enhanced binding of GATA-6 to the GATA motif of the CAV1 promoter in BSM from mice with surgically induced PBOO and in men with BPH-induced PBOO. Next tested was whether expression of GATA-6 itself is regulated under the same conditions. GATA-6 protein and mRNA were measured at immunoblotting and RT-PCR, respectively, in healthy and PBOO or BPH bladder samples. Over 14 days, GATA-6 mRNA expression increased in PBOO bladders relative to GAPDH expression in mice (Figure 7, A and B), and GATA-6 protein similarly increased during the same period (Figure 7, C and D). Likewise, GATA-6 mRNA and protein levels were increased in bladder samples from patients with BPH compared with healthy control individuals (Figure 8, A–D). Immunofluoresence imaging analysis demonstrated increased expression of GATA-6 in bladder smooth muscle from patients with BPH compared with healthy bladders (Figure 8E). These results suggest that PBOO-induced remodeling of BSM alters GATA-6 mRNA and protein expression in both mice and human BSM.
Figure 7.
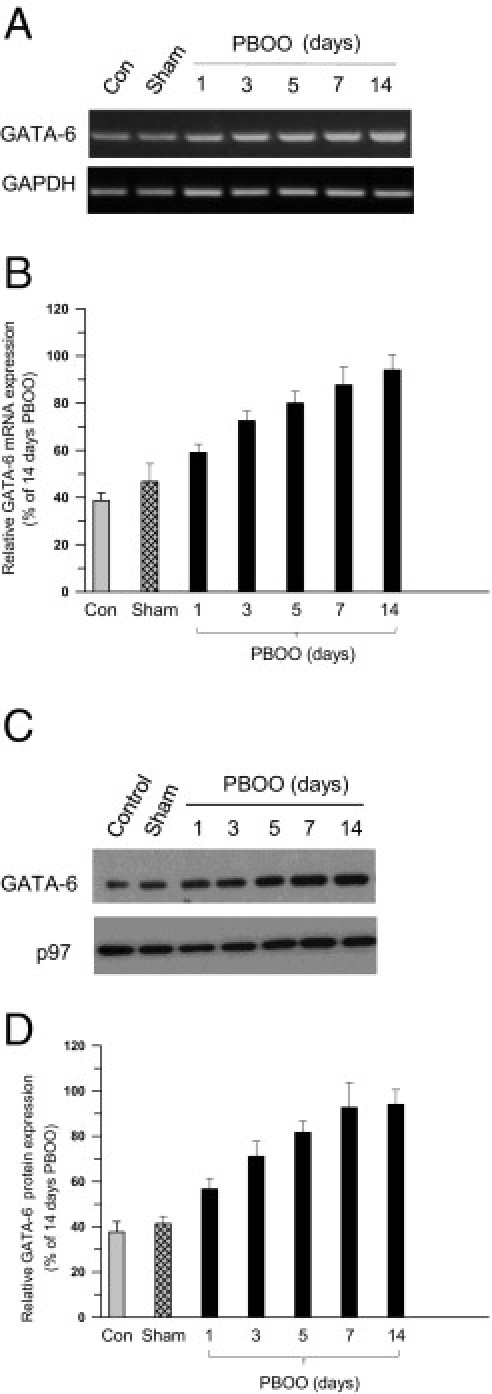
Increased expression of GATA-6 in obstruction-induced BSM hypertrophy in mice. A: RNA was prepared from control, sham, and PBOO mouse BSM tissues and subjected to RT-PCR for GATA-6 and GAPDH as control. B: Densitometric analysis of bands from A presented as GATA-6 expression (upper panel) relative to GAPDH (lower panel). C: Nuclear protein was extracted from the same samples as in A and subjected to immunoblot analysis for anti–GATA-6 (upper panel) and nuclear protein p97 (lower panel) as control. D: Densitometric analysis of bands from C reported as GATA-6 expression relative to p97. In B and D, data are given as mean (SD) from four independent experiments.
Figure 8.
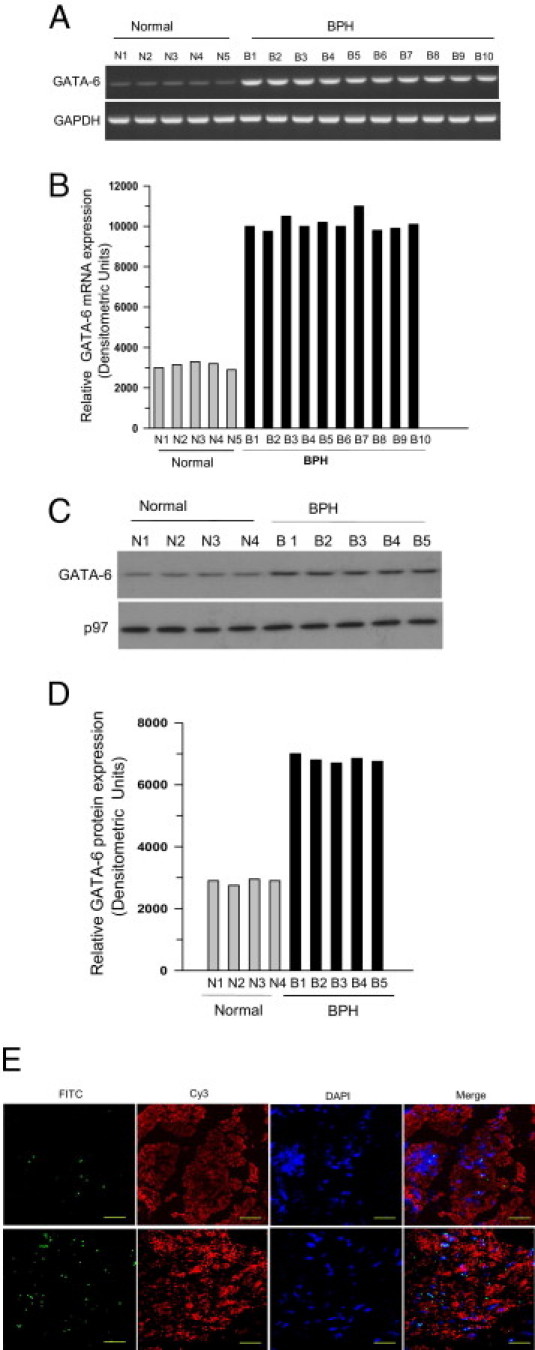
Increased expression of GATA-6 in obstruction-induced BSM hypertrophy in men with BPH. A: RNA was extracted from healthy (N) and BPH (B) human BSM tissues and subjected to RT-PCR for GATA-6 (upper panel) and GAPDH (lower panel) as control. B: Densitometric analysis of bands from A reported as GATA-6 expression relative to GAPDH. C: Nuclear protein was extracted from the same samples and subjected to immunoblotting. D: Densitometric analysis of bands in C presented as GATA-6 (upper panel in C) expression relative to nuclear protein p97 (lower panel in C). Data are from individual patients. E: Immunofluorescence images of endogenous GATA-6 in human BSM from healthy men (upper panel) and patients with BPH (lower panel). Human BSM tissue was stained with anti-GATA-6 polyclonal antibody (1:100) followed by FITC-conjugated secondary antibody. The smooth muscle tissues were also stained with Cy3-conjugated smooth muscle myosin heavy-chain 1. Slides were mounted in VectaShield medium containing DAPI to counterstain nuclei. Original magnification, ×63. Scale bars = 20 μm.
GATA-5 Expression Level Is Not Altered in Mouse PBOO and Human BPH Bladder Samples
Promoter pull-down analysis revealed that transcription factor GATA-5 does not interact with murine CAV1 promoter. Expression of GATA-5 was profiled in smooth muscle tissue isolated from control and PBOO or BPH bladder samples. GATA-5 protein and mRNA were measured at immunoblotting and RT-PCR, respectively, in healthy and PBOO or BPH bladder samples. Over 14 days, GATA-5 mRNA expression did not change in PBOO bladders relative to GAPDH expression in mice (Figure 9A), and GATA-5 protein similarly did not change during the same period (Figure 9B). Likewise, GATA-5 mRNA and protein levels were not changed in bladder samples from patients with BPH compared with those from healthy control individuals (Figure 9, C and D).
Figure 9.
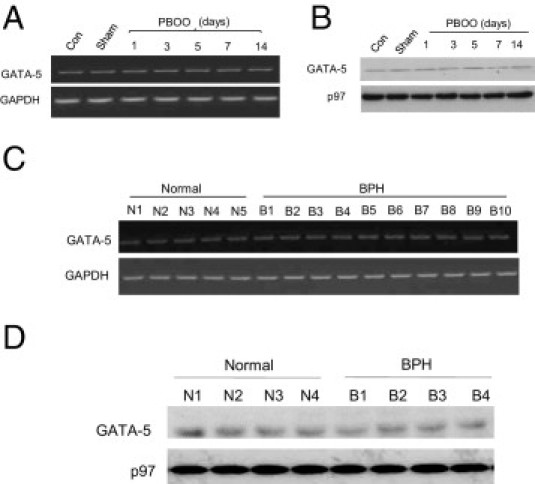
GATA-5 profiling in normal and obstructed hypertrophic BSM in mice and human beings. A: RNA was prepared from control, sham, and PBOO mouse bladder smooth muscle and subjected to RT-PCR for GATA-5 and GAPDH as control. B: Nuclear protein was extracted from the same samples and subjected to immunoblotting for anti-GATA-5 (upper panel) and nuclear protein p97 (lower panel) as control. C: RNA was extracted from healthy (N) and BPH (B) human BSM and subjected to RT-PCR for GATA-5 (upper panel) and GAPDH (lower panel) as control. D: Nuclear protein was extracted from the same samples and subjected to immunoblotting for anti-GATA-5 (upper panel) and nuclear protein p97 (lower panel) as control.
Transcriptional Repression of the CAV1 Gene by GATA-6
Because GATA-6 binds to its consensus sequence in the CAV1 promoter in PBOO, the effect of GATA-6 overexpression in regulating CAV1 gene expression was examined. GATA-6 cDNA was introduced into human bladder primary SMCs via transduction with adenovirus encoding LacZ or GATA-6. At 72 hours after infection, cells were harvested, and endogenous protein and mRNA expression were analyzed at immunoblotting and RT-PCR, respectively. It was observed that overexpression of GATA-6 down-regulated endogenous caveolin-1 mRNA (Figure 10, A and B) and protein (Figure 10, C and D) expression relative to LacZ in human BSM cells. Similar results were obtained using mouse bladder SMCs (data not shown), confirming the consistency of this observation.
Figure 10.
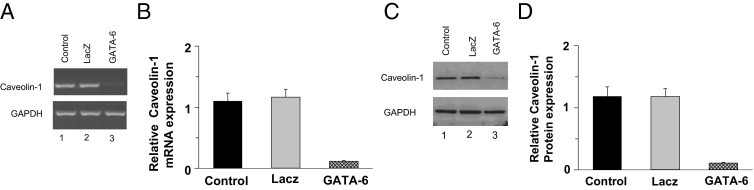
GATA-6 negatively regulates caveolin-1 (CAV1) gene expression in primary BSM cells. A: Human BSM cells were transduced with adenovirus encoding GATA-6 or LacZ for 72 hours. RNA was extracted and subjected to RT-PCR analysis for caveolin-1 and GAPDH. B: Densitometric analysis of bands from A presented as caveolin-1 expression relative to GAPDH. C: Protein extracts were prepared from the same samples as in A and were subjected to immunoblotting for caveolin-1 and GAPDH. D: Densitometric analysis of bands from C presented as caveolin-1 expression relative to GAPDH. In B and D, data are given as mean (SD) from four independent experiments.
The specific role of GATA-6 in regulating the transcriptional activity of the CAV1 promoter was studied via co-transfection of promoter constructs containing the GATA-6 binding motif upstream of the luciferase gene (Figure 11A) to human and mouse primary BSM cells. As expected, mutation of the GATA-6-binding consensus core nucleotides to CCCC (MUT) presumably disrupted GATA-6 recruitment and thereby drove transcription of the construct to result in greater luciferase activity, as compared with cells transfected with the construct containing the WT GATA-6–binding motif (Figure 11B). The repressive activity of GATA-6 on luciferase expression was almost completely reversed by co-expression of the mutant GATA-6–binding motif luciferase plasmid with GATA-6 cDNA, which is in full agreement with previous studies that demonstrated that core nucleotides of the GATA-6 consensus motif are important for GATA-6 binding.16–18 However, mutations in the outside region of the GATA-6 binding site, which did not impair GATA-6 recruitment, resulted in promoter activity similar to WT (data not shown).
Figure 11.
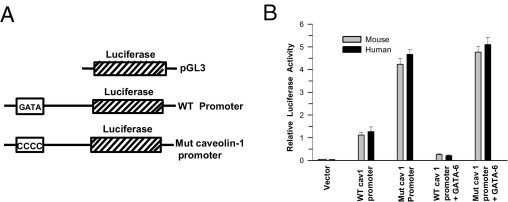
Mutation of the GATA motif in the caveolin-1 (CAV1) promoter increases luciferase activity in transfected BSM cells. A: Schema of the luciferase constructs. The GATA motif and its mutated sequence of the CAV1 promoter are indicated. B: Murine and human BSM cells were transfected with murine (gray bars) or human (black bars) CAV1 promoter, as described in A, and with GATA-6 cDNA when indicated. Luciferase activity was measured in each sample after 72 hours and is presented relative to that of Renilla luciferase. Data are given as mean (SD) from five independent experiments.
GATA6 Gene Silencing Up-Regulates CAV1 Gene Transcription and Promoter Activity in BSM Cells
The role of GATA6 in the transcriptional regulation of CAV1 gene expression was further investigated using a lentiviral vector containing GATA6 siRNA, which enables gene silencing in both dividing and nondividing cells.47 This has been successfully used for GATA6 gene silencing in human primary BSM cells.22 Thus, this system was used to suppress endogenous GATA6 expression in human primary BSM cells. RT-PCR and immunoblot analysis were used to determine the steady-state level of GATA-6 in shRNA-transfected human BSM cells. As expected, both mRNA (Figure 12, A and B) and protein (Figure 12, C and D) GATA-6 levels in BSM cells transfected with shRNA specific for GATA-6 were significantly reduced compared with nonspecific (scrambled) siRNA-transfected cells. As before, identical results were obtained using murine BSM cells (data not shown).
Figure 12.
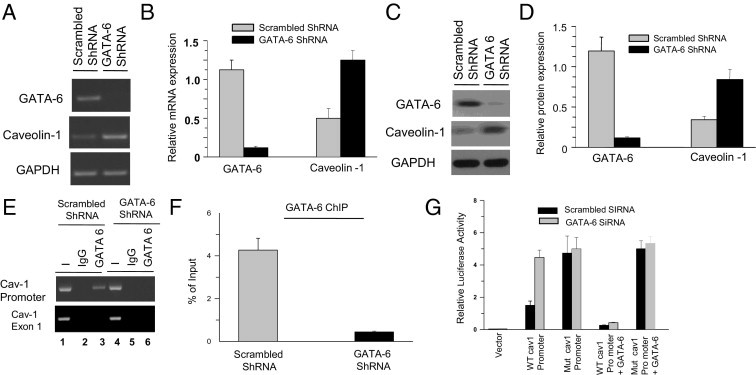
GATA-6 (GATA6) silencing affects caveolin-1 (CAV1) gene expression, transcriptional activity, and promoter occupancy in primary human BSM cells. A: RT-PCR analysis of GATA-6 and caveolin-1 in scrambled and GATA-6 shRNA-transfected human BSM cells. GAPDH was amplified as control. B: Densitometric analysis of the bands in A for GATA-6 and caveolin mRNA is presented relative to GAPDH. C: Immunoblot analysis of total protein from cells transduced with scrambled and GATA-6 shRNA lentivirus vectors. GAPDH was probed as a loading control. D: Densitometric analysis of the bands in C for GATA-6 and caveolin-1 protein relative to GAPDH. E: ChIP analysis of the CAV1 promoter with GATA-6 antibody in human BSM cells transduced with scrambled and GATA-6 shRNA. GATA-6 binding motif on human CAV1 promoter was amplified at PCR using the primers given in Table 1. Preimmune serum was used as negative control. F: Quantitation of the band intensity from E is expressed as percentage of input DNA. G: Relative luciferase activity in human BSM cells transfected with scrambled (black bars) and GATA-6 (gray bars) siRNA. Reporter firefly luciferase activities were normalized to Renilla luciferase activity. In B, D, F, and G data are given as mean (SD) of three independent experiments.
ChIP experiments were performed using human BSM cells transfected with scrambled or GATA-6 shRNA. Samples immunoprecipitated with anti–GATA-6 antibody from human (Figure 12E, lane 3) scrambled shRNA-transfected cells yielded prominent PCR products, whereas GATA-6 shRNA-transfected cells failed to amplify any detectable DNA (Figure 12, E and F). Similarly, samples immunoprecipitated with preimmune rabbit IgG from scrambled and GATA-6 shRNA-transfected cells did not amplify any PCR products (Figure 12E, lanes 2 and 5). Transfection of GATA-6 shRNA in these cells resulted in loss of GATA-6 binding to the CAV1 promoter (Figure 12E, lane 6), and exon-1 was amplified as negative control (Figure 12E). The relative binding of GATA-6 to the GATA motif on the CAV1 promoter is presented as percentage of input chromatin for human BSM cells (Figure 12F). Again, identical results were obtained using murine BSM cells (data not shown). Together, these results suggest that silencing of GATA-6 impairs its association with its cognate DNA sequence on the CAV1 promoter, both in human and mouse species.
To examine the effect of endogenous GATA6 gene silencing in vivo in regulating CAV1 promoter activity, GATA-6 siRNA oligonucleotides were co-transfected with CAV1 promoter in human primary BSM cells (Figure 12G) via electroporation. Promoter analysis using human CAV1 promoter constructs did not demonstrate any significant effects on luciferase expression after transfection with nonspecific siRNA. However, luciferase activity driven by WT human CAV1 promoters was fourfold higher in GATA-6 knockdown cells compared with scrambled siRNA-transfected cells, which was reversed by addition of GATA-6 cDNA. These results suggest that repression of CAV1 gene expression is due to selective effects of GATA-6 on the CAV1 promoter, and provide direct proof that GATA-6 participates in transcriptional repression of CAV1 gene expression in human and murine BSM cells (data not shown).
GATA5 Gene Silencing Does Not Affect CAV1 Gene Transcription and Promoter Activity in BSM Cells
Effect of GATA5 gene silencing on the transcriptional regulation of CAV1 gene expression was further investigated using GATA-5 shRNA. RT-PCR and immunoblot analysis were used to determine the steady-state level of GATA-5 in shRNA-transfected human BSM cells. GATA-5 levels in BSM cells transfected with shRNA specific for GATA-5 were significantly reduced compared with nonspecific (scrambled) shRNA-transfected cells (Figure 13, A–D). However, GATA5 gene knockdown did not alter the caveolin-1 mRNA and protein level in human BSM cells (Figure 13, A–D). Identical results were obtained using murine BSM cells (data not shown). To examine the effect of endogenous GATA5 gene silencing in vivo in regulating CAV1 promoter activity, GATA-5 shRNA plasmid was co-transfected with CAV1 promoter in human primary BSM cells (Figure 13E) via electroporation. Promoter analysis using human and murine CAV1 promoter constructs did not demonstrate any significant effects on luciferase expression after transfection with nonspecific and GATA-5 shRNA. Identical results were obtained using murine BSM cells (data not shown). These results suggest that the transcription factor GATA-5 does not have any effect on CAV1 gene expression in human and murine BSM.
Figure 13.
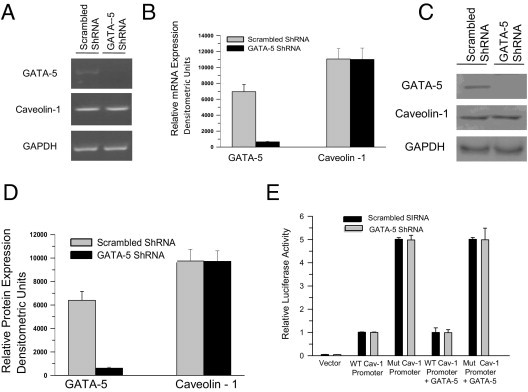
GATA-5 (GATA5) silencing did not affect caveolin-1 (CAV1) gene expression and transcriptional activity in human BSM cells. A: RT-PCR analysis of GATA-5 and caveolin-1 in scrambled and GATA-5 shRNA-transfected human BSM cells. GAPDH was amplified as control. B: Densitometry analysis of the bands in A for GATA-5 and caveolin-1 mRNA is presented relative to GAPDH. C: Immunoblot analysis of total protein from cells transfected with scrambled and GATA-5 shRNA plasmid. GAPDH was probed as a loading control. D: Densitometry analysis of the bands in C for GATA-5 and caveolin-1 protein relative to GAPDH. E: Relative luciferase activity in human BSM cells transfected with scrambled (black bars) and GATA-5 (gray bars) shRNA plasmid. Reporter firefly luciferase activities were normalized to Renilla luciferase activity. In B and D, data are given as mean (SD) of three independent experiments.
Discussion
The present study described the molecular mechanism of transcriptional regulation of CAV1 gene expression in BSM, thereby unraveling the process that mediates loss of CAV1 gene expression during PBOO-induced BSM remodeling and hypertrophy in the mouse model of PBOO and in men with BPH. It was demonstrated that the transcription factor GATA-6 acts as a repressor of CAV1 gene expression and that this repression in BSM in response to bladder outlet obstruction is mediated by overexpression of GATA-6.
The loss of caveolin-1 expression in BSM from BPH-induced outlet obstruction in men and in mouse PBOO bladders is parallel with overexpression of GATA-6 in the same tissues. PBOO-induced overexpression of GATA-6 in BSM correlates with increased binding of GATA-6 to its consensus DNA binding motif on the CAV1 promoter compared with its binding to the GATA motif from sham bladders, as evidenced at ChIP analysis and in promoter pull-down experiments. Results from experiments using cultured BSM cells and intact BSM tissue from mice and human patients suggest that transcription factor GATA-6 acts as a transcriptional repressor. Being a transcriptional repressor of both human and murine CAV1 gene transcription, GATA-6 suppresses CAV1 gene expression in obstructed BSM, likely as a result of its increased expression in the these tissues.
An earlier study of the regulation of CAV1 gene transcription in human skin fibroblasts involved the complex of SP-1, E2F/DP-1, and p53 in response to cholesterol homeostasis.48 CAV1 is transcriptionally induced in colon cancer cells in response to conditional expression of a full-length adenomatous polyposis coli (APC) gene. The induction of caveolin-1 by APC is mediated by both FOXO1a, a member of the forkhead box family of transcription factors, and c-myc.49 CAV1 expression is regulated by the ETS family of transcription factors through an ETS-binding DNA motif on the caveolin-1 promoter in lung epithelial cells.50 An earlier study suggested that an ETS family transcription factor, Net, may suppress CAV1 transcription in primary lung tumors, and PEA3 may activate CAV1 transcription in metastatic lymph nodes.51
In bladder cancer, caveolin-1 expression is undetectable in benign urothelium, higher in urothelial carcinomas, and maximal in tumor cells.52 Caveolin-1 expression is correlated with tumor stage and grade of bladder transitional cell carcinoma (TCC).53,54Caveolin-1 expression was correlated significantly with T stage and grade of bladder TCC, which strongly indicates that caveolin-1 is involved in tumor progression and that the increased expression of caveolin-1 is a late event in urothelial carcinomas.55 The increased expression of caveolin-1 is associated with tumor progression and poor prognosis in upper urinary tract TCC, which suggests that caveolin-1 may have an important role in progression of upper urinary tract TCC.55 Hypermethylation of the caveolin-1 gene and abnormal caveolin-1 protein expression have crucial roles in cell differentiation and in the phonotypical conversion of TCC into nonurothelial carcinomas.56
Among the transcription factors that were studied at promoter pull-down analysis with the nuclear extract from murine sham and PBOO BSM tissues, transcription factor Ets-2 is overexpressed in human bladder cancer,57 and SP-1 regulates tumor necrosis factor-α–induced MMP-9 expression through ERK1/2 and p38 MAP kinase in bladder cancer cells.58 However, in murine and human obstructed BSM tissues, Ets-2 and SP-1 are not overexpressed. Moreover, E2F and Sp-1 did not interact with murine CAV1 promoter as revealed at promoter pull-down analysis. The epithelial cells were removed from the murine and human bladder, and only the smooth muscle tissues were used for the experiment.
Using multiple approaches, the present study demonstrated for the first time, to our knowledge, that GATA-6 acts as a repressor of CAV1 gene transcription in BSM and that this repression in BSM from obstructed bladders is mediated by GATA-6 overexpression. An important finding of the study is involvement of GATA-6 in transcriptional regulation of CAV1 gene expression in pathologic smooth muscle from dysfunctional bladders, relating the changes in the molecular regulation of CAV1 to organ function or dysfunction.
Gain-of-function and loss-of-function approaches in experiments with cultured BSM cells suggest a role for GATA-6 in transcriptional suppression of CAV1 gene expression. Down-regulation of GATA-6 protein levels in primary BSM cells by RNA interference led to increased caveolin-1 mRNA and protein production. In contrast, overexpression of GATA-6 in murine and human BSM cells repressed CAV1 gene expression, which suggests an inverse relationship between expression of GATA-6 and caveolin-1.
ChIP assay data demonstrated that GATA-6 is recruited to the CAV1 promoter. GATA6 gene silencing experiments in cultured BSM cells failed to detect the GATA motif on the CAV1 promoter in the ChIP assays, substantiating its role in transcriptional repression. The CAV1 promoter reporter constructs in transient transfection studies demonstrated the requirement of the GATA-6 binding site (GATA motif) for promoter activity, as evidenced by the lack of GATA-6–mediated suppression of promoter activity on point mutation of a putative GATA binding motif within the caveolin-1 proximal promoter region. These data confirm that GATA-6 binding to this site is required for CAV1 gene repression in SMCs. Although GATA-5 is expressed in the bladder wall,59 the expression level of this protein did not change in PBOO or BPH bladder samples compared with control, and GATA5 gene silencing in human and murine bladder primary smooth muscle did not affect CAV1 gene expression and promoter activity.
To our knowledge, this is the first report to demonstrate differential GATA-6 expression between diseased and healthy BSM tissues and the involvement of GATA-6 in transcriptional repression of caveolin-1 gene expression. Caveolin-1 is the most important member of a family of membrane proteins that are the major coat proteins of caveolae. This study links transcriptional repression of the CAV1 gene with GATA-6, which is involved in smooth muscle differentiation. Alteration in the caveolae, which harbor several proteins involved in signal transduction in smooth muscle, would likely contribute to changes in the signaling that regulates smooth muscle contraction.
In addition to its relevance to regulation of this gene in smooth muscle, this observation also has implications in several physiologic and pathologic conditions including cardiac hypertrophy, fibrosis,60 and tumor cell metastasis,61,62 and the pathologic features of these diseases include loss of CAV1 gene expression. Consistent with these findings, it was demonstrated that there is loss of caveolin-1 in human BSM in response to BPH-induced bladder outlet obstruction, and these findings were reproduced in a mouse model via surgical ligation of the urethra to induce bladder outlet obstruction. Bladder outlet obstruction–induced loss of caveolin-1 in smooth muscle could be a contributing factor in impaired smooth muscle contraction observed in murine PBOO bladders. Since GATA-6 is not expressed in smooth muscle from all tissues, demonstrating robust expression in the vasculature, BSM, and cardiac cells, it is tempting to speculate that recruitment of GATA-6 to the CAV1 promoter could elicit tissue- and context-specific regulation of CAV1 expression in smooth and cardiac muscle cells.
The increasing interest in caveolae stems primarily from the fact that these structures seem to sequester and regulate a variety of signaling molecules7 including receptors, kinases, and phospholipids. G protein–coupled receptors and tyrosine kinase receptors are believed to cluster in caveolae, and the exciting possibility that caveolae could provide a platform for interactions between the sarcoplasmic reticulum and plasmalemmal ion channels in smooth muscle is emerging.63 Moreover, messengers involved in Ca2+ sensitization of myosin phosphorylation, an important regulatory pathway in maintenance of smooth muscle tone, may depend on caveolae or caveolin. Mice with a disrupted CAV1 gene exhibit pronounced cardiovascular phenotypes including cardiomyopathy, severe pulmonary hypertension, and impaired BSM contraction.60,30 Caveolae seem to constitute an important signaling domain that has a role not only in regulation of smooth muscle tone64 but also in proliferation such as observed in neointima formation and atherosclerosis.63 Deletion of CAV1 leads to significant bladder dysfunction and urogenital organ changes in aged male mice.30
In summary, the present study demonstrated that GATA-6 is a transcriptional repressor of both the human and murine CAV1 genes in BSM. In addition, a new mode of caveolin-1 gene regulation has been established in which the GATA motif on the CAV1 promoter mediates its cognate gene expression through GATA-6 recruitment to the promoter in these cells. Together, these observations provide a starting point for future studies with the objective of understanding the GATA-6–dependent signaling under pathologic conditions in addition to its function in smooth muscle differentiation and maintenance. Data from the present study provide a rationale for evaluating therapeutic strategies targeting GATA-6–mediated transcriptional regulation of CAV1 gene expression and smooth muscle contractility in conditions associated with decreased contractility including that observed in BPH-induced bladder dysfunction. Currently available drugs for improving detrusor contractility seem to have limited efficacy and adverse effects that would limit their use at high dosages. Both limitations may be improved by targeting the GATA-6–mediated molecular pathway that is up-regulated in BPH-induced bladder dysfunction in men.
Acknowledgment
We thank Dr. Gomes (University of São Paolo, São Paolo, Brazil) for frozen bladder tissue samples (collected using IRB protocol No. 811/04 at the University of São Paulo); Dr. Hasmeena Kathuria (Boston University School of Medicine, Boston, MA) for donating the luciferase reporter plasmids containing the mouse and human CAV1 promoters; and Ms. Jocelyn McCabe for assistance in preparing the manuscript.
Footnotes
Supported by grant P50 DK052620 from the George O'Brien Urology Research Center.
References
- 1.Wein A.J. Pathophysiology and categorization of voiding dysfunction: Campbell's Urology. In: Walsh P.C., Retik A.B., Vaughan D.E., Wein A.J., editors. Saunders; Philadelphia, PA: 1998. pp. 917–926. [Google Scholar]
- 2.Abrams P., Donovan J.L., de la Rosette J.J., Schäfer W. International Continence Society “Benign Prostatic Hyperplasia” Study: background, aims, and methodology. Neurourol Urodyn. 1997;16:79–91. [PubMed] [Google Scholar]
- 3.DiSanto M.E., Stein R., Chang S., Hypolite J.A., Zheng Y., Zderic S., Wein A.J., Chacko S. Alteration in the expression of myosin isoforms in detrusor smooth muscle following bladder outlet obstruction. Am J Physiol Cell Physiol. 2003;285:C1397–C1410. doi: 10.1152/ajpcell.00513.2002. [DOI] [PubMed] [Google Scholar]
- 4.Bing W., Chang S., Hypolite J.A., DiSanto M.E., Zderic S.A., Rolf L., Wein A.J., Chacko S. Obstruction-induced changes in urinary bladder smooth muscle contractility: a role for Rho kinase. Am J Physiol Renal Physiol. 2003;285:F990–F997. doi: 10.1152/ajprenal.00378.2002. [DOI] [PubMed] [Google Scholar]
- 5.Levin R.M., Haugaard N., O'Connor L., Buttyan R., Das A., Dixon J.S., Gosling J.A. Obstructive response of human bladder to BPH vs. rabbit bladder response to partial outlet obstruction: a direct comparison. Neurourol Urodyn. 2000;19:609–629. doi: 10.1002/1520-6777(2000)19:5<609::aid-nau7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 6.Polyák E., Boopathi E., Mohanan S., Deng M., Zderic S.A., Wein A.J., Chacko S. Alterations in caveolin expression and ultrastructure after bladder smooth muscle hypertrophy. J Urol. 2009;182:2497–2503. doi: 10.1016/j.juro.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Galbiati F., Razani B Lisanti M.P. Emerging themes in lipid rafts and caveolae. Cell. 2001;106:403–411. doi: 10.1016/s0092-8674(01)00472-x. [DOI] [PubMed] [Google Scholar]
- 8.Fielding C.J., Fielding P.E. Relationship between cholesterol trafficking and signaling in rafts and caveolae. Biochim Biophys Acta (BBA)–Biomembranes. 2003;1610:219–228. doi: 10.1016/s0005-2736(03)00020-8. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto T., Schlege A., Scherer P.E., Lisanti M.P. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 10.Razani B., Lisanti M.P. Caveolin-deficient mice: insights into caveolar function human disease. J Clin Invest. 2001;108:1553–1561. doi: 10.1172/JCI14611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Couet S., Li T., Okamoto T., Ikezu, Lisanti M.P. Identification of peptide and protein ligands for the caveolin-scaffolding domain: implications for the interaction of caveolin with caveolae-associated proteins. J Biol Chem. 1997;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- 12.Bist A., Fielding P.E., Fielding C.J. Two sterol regulatory element-like sequences mediate up-regulation of caveolin gene transcription in response to low density lipoprotein free cholesterol. Proc Natl Acad Sci USA. 1997;94:10693–10698. doi: 10.1073/pnas.94.20.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Heuvel A.P., Schulze A., Burgering B.M. Direct control of caveolin-1 expression by FOXO transcription factors. Biochem J. 2005;385:795–802. doi: 10.1042/BJ20041449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molkentin J.D. The zinc finger–containing transcription factors GATA-4, -5, and -6: ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 15.Burch J.B. Regulation of GATA gene expression during vertebrate development. Semin Cell Dev Bio. 2005;16:71–81. doi: 10.1016/j.semcdb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Orkin S.H. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 17.Ko L.J., Engel J.D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai Y., Nakagawa R., Sato R., Maeda M. Selection of DNA binding sites for human transcriptional regulator GATA-6. Biochem Biophys Res Commun. 1998;250:682–688. doi: 10.1006/bbrc.1998.9374. [DOI] [PubMed] [Google Scholar]
- 19.Chang D.F., Belaguli N.S., Iyer D., Roberts W.B., Wu S.P., Dong X.R., Marx J.G., Moore M.S., Beckerle M.C., Majesky M.W., Schwartz R.J. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4:107–118. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 20.Yin F., Herring B.P. GATA-6 can act as a positive or negative regulator of smooth muscle-specific gene expression. J Biol Chem. 2005;280:4745–4752. doi: 10.1074/jbc.M411585200. [DOI] [PubMed] [Google Scholar]
- 21.Shirvani S., Xiang F., Koibuchi N., Chin M.T. CHF1/Hey2 suppresses SM-MHC promoter activity through an interaction with GATA-6. Biochem Biophys Res Commun. 2006;339:151–156. doi: 10.1016/j.bbrc.2005.10.190. [DOI] [PubMed] [Google Scholar]
- 22.Kanematsu A., Ramachandran A., Adam R.M. GATA-6 mediates human bladder smooth muscle differentiation: involvement of a novel enhancer element in regulating alpha-smooth muscle actin gene expression. Am J Physiol Cell Physiol. 2007;293:C1093–C1102. doi: 10.1152/ajpcell.00225.2007. [DOI] [PubMed] [Google Scholar]
- 23.Lepore J.J., Cappola T.P., Mericko P.A., Morrisey E.E., Parmacek M.S. GATA-6 regulates genes promoting synthetic functions in vascular SMCs. Arterioscle Thromb Vasc Biol. 2005;25:309–314. doi: 10.1161/01.ATV.0000152725.76020.3c. [DOI] [PubMed] [Google Scholar]
- 24.Liang Q., De Windt L.J., Witt S.A., Kimball T.R., Markham B.E., Molkentin J.D. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J Biol Chem. 2001;276:30245–30253. doi: 10.1074/jbc.M102174200. [DOI] [PubMed] [Google Scholar]
- 25.Morrisey E.E., Ip H.S., Lu M.M., Parmacek M.S. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- 26.Patel H.H., Zhang S., Murray F., Suda R.Y., Head B.P., Yokoyama U., Swaney J.S., Niesman I.R., Schermuly R.T., Pullamsetti S.S., Thistlethwaite P.A., Miyanohara A., Farquhar M.G., Yuan J.X., Insel P.A. Increased smooth muscle cell expression of caveolin-1 and caveolae contribute to the pathophysiology of idiopathic pulmonary arterial hypertension. FASEB J. 2007;11:2970–2979. doi: 10.1096/fj.07-8424com. [DOI] [PubMed] [Google Scholar]
- 27.Lipardi C., Mora R., Colomer V., Paladino S., Nitsch L., Rodriguez-Boulan E., Zurzolo C. Caveolin transfection results in caveolae formation but not apical sorting of glycosylphosphatidylinositol (GPI)–anchored proteins in epithelial cells. J Cell Biol. 1998;140:617–626. doi: 10.1083/jcb.140.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stehr M., Adam R.M., Khoury J., Zhuang L., Solomon K.R., Peters C.A., Freeman M.R. Platelet derived growth factor–BB is a potent mitogen for rat ureteral and human bladder SMCs: dependence on lipid rafts for cell signaling. J Urol. 2003;169:1165–1170. doi: 10.1097/01.ju.0000041501.01323.b9. [DOI] [PubMed] [Google Scholar]
- 29.Stehr M., Estrada C.R., Khoury J., Danciu T.E., Sullivan M.P., Peters C.A., Solomon K.R., Freeman M.R., Adam R.M. Caveolae are negative regulators of transforming growth factor-beta1 signaling in ureteral SMCs. J Urol. 2004;172:2451–2455. doi: 10.1097/01.ju.0000138084.53577.ca. [DOI] [PubMed] [Google Scholar]
- 30.Woodman S.E., Cheung M.W., Tarr M., North A.C., Schubert W., Lagaud G., Marks C.B., Russell R.G., Hassan G.S., Factor S.M., Christ G.J., Lisanti M.P. Urogenital alterations in aged male caveolin-1 knockout mice. J Urol. 2004;171:950–957. doi: 10.1097/01.ju.0000105102.72295.b8. [DOI] [PubMed] [Google Scholar]
- 31.Cao G., Yang G., Timme T.L., Saika T., Truong L.D., Satoh T., Goltsov A., Park S.H., Men T., Kusaka N., Tian W., Ren C., Wang H., Kadmon D., Cai W.W., Chinault A.C., Boone T.B., Bradley A., Thompson T.C. Disruption of the caveolin-1 gene impairs renal calcium reabsorption and leads to hypercalciuria and urolithiasis. Am J Pathol. 2003;162:1241–1248. doi: 10.1016/S0002-9440(10)63920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koleske A.J., Baltimore D., Lisanti M.P. Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci USA. 1995;92:1381–1385. doi: 10.1073/pnas.92.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drab M., Verkade P., Elger M., Kasper M., Lohn M., Lauterbach B., Menne J., Lindschau C., Mende F., Luft F.C., Schedl A., Haller H., Kurzchalia T.V. Loss of caveolae: vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- 34.Rahman A., Swärd K. The role of caveolin-1 in cardiovascular regulation. Acta Physiol (Oxf) 2009;195:231–245. doi: 10.1111/j.1748-1716.2008.01907.x. [DOI] [PubMed] [Google Scholar]
- 35.Austin J.C., Chacko S.K., DiSanto M., Canning D.A., Zderic S.A. A male murine model of partial bladder outlet obstruction reveals changes in detrusor morphology, contractility and myosin isoform expression. J Urol. 2004;172:1524–1528. doi: 10.1097/01.ju.0000138045.61378.96. [DOI] [PubMed] [Google Scholar]
- 36.Abrams P. Bladder outlet obstruction index, bladder contractility index and bladder voiding efficiency: three simple indices to define bladder voiding function. BJU Int. 1999;84:14–15. doi: 10.1046/j.1464-410x.1999.00121.x. [DOI] [PubMed] [Google Scholar]
- 37.Abrams P., Cardozo L., Fall M., Griffiths D., Rosier P., Ulmsten U., van Kerrebroeck P., Victor A., Wein A. The standardization of terminology of lower urinary tract function: report from the Standardization Sub-Committee of the International Continence Society. Neurourol Urodyn. 2002;21:167–178. doi: 10.1002/nau.10052. [DOI] [PubMed] [Google Scholar]
- 38.Griffiths D., Hofner K., van M.R., Rollema H.J., Spangberg A., Gleason D. Standardization of terminology of lower urinary tract function: pressure-flow studies of voiding, urethral resistance, and urethral obstruction: International Continence Society Subcommittee on Standardization of Terminology of Pressure-Flow Studies. Neurourol Urodyn. 1997;16:1–18. doi: 10.1002/(sici)1520-6777(1997)16:1<1::aid-nau1>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber E., Matthias P., Muller M.M. Schaffner W: Rapid detection of octamer binding proteins with “mini-extracts” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419–6424. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang E., Stein R., Chang S., Zheng Y., Zderic S.A., Wein A.J., Chacko S. Smooth muscle hypertrophy following partial bladder outlet obstruction is associated with overexpression of non-muscle caldesmon. Am J Pathol. 2000;164:601–612. doi: 10.1016/S0002-9440(10)63149-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masternak K.A., Muhlethaler-Mottet J., Villard M., Zufferey V., Steimle R., Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 42.Kadonaga J.T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Neill L.P., Turner B.M. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okada T., Ramsey W.J., Munir J., Wildner O., Blaese R.M. Efficient directional cloning of recombinant adenovirus vectors using DNA-protein complex. Nucleic Acids Res. 1998;26:1947–1950. doi: 10.1093/nar/26.8.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. Matlnspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [Epub ahead of press April 28, 2005] [DOI] [PubMed] [Google Scholar]
- 47.Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S., Sabatini D.M., Chen I.S., Hahn W.C., Sharp P.A., Weinberg R.A., Novina C.D. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bist J., Fielding C., Fielding P.E. p53 Regulates caveolin gene transcription, cell cholesterol, and growth by a novel mechanism. Biochemistry. 2000;39:1966–1972. doi: 10.1021/bi991721h. [DOI] [PubMed] [Google Scholar]
- 49.Roy U.K., Henkhaus R.S., Ignatenko N.A., Mora M., Fultz K.E., Gerner E.W. Wild-type APC regulates caveolin-1 expression in human colon adenocarcinoma cell lines via FOXO1a and C-myc. Mol Carcinog. 2008;47:947–955. doi: 10.1002/mc.20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kathuria H., Cao Y.X., Ramirez M.I., Williams M.C. Transcription of the caveolin-1 gene is differentially regulated in lung type I epithelial and endothelial cell lines: a role for ETS proteins in epithelial cell expression. J Biol Chem. 2004;279:30028–30036. doi: 10.1074/jbc.M402236200. [DOI] [PubMed] [Google Scholar]
- 51.Sloan K.A., Marquez H.A., Li J., Cao Y., Hinds A., O'Hara C.J., Kathuria S., Ramirez M.I., Williams M.C., Kathuria H. Increased PEA3/E1AF and decreased Net/Elk-3, both ETS proteins, characterize human NSCLC progression and regulate caveolin-1 transcription in Calu-1 and NCI-H23 NSCLC cell lines. Carcinogenesis. 2009;30:1433–1442. doi: 10.1093/carcin/bgp129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fong A., Garcia E., Gwynn L., Lisanti M.P., Fazzari M.J., Li M. Expression of caveolin-1 and caveolin-2 in urothelial carcinoma of the urinary bladder correlates with tumor grade and squamous differentiation. Am J Clin Pathol. 2003;120:93–100. doi: 10.1309/292N-HAYN-WAVR-EJ37. [DOI] [PubMed] [Google Scholar]
- 53.Rajjayabun P.H., Garg S., Durkan G.C., Charlton R., Robinson M.C., Mellon J.K. Caveolin-1 expression is associated with high-grade bladder cancer. Urology. 2001;58:811–814. doi: 10.1016/s0090-4295(01)01337-1. [DOI] [PubMed] [Google Scholar]
- 54.Thomas S., Overdevest J.B., Nitz M.D., Williams P.D., Owens C.R., Sanchez-Carbayo M., Frierson H.F., Jr, Schwartz M.A., Theodorescu D. Src and caveolin-1 reciprocally regulate metastasis via a common downstream signaling pathway in bladder cancer. Cancer Res. 2010;71:832–841. doi: 10.1158/0008-5472.CAN-10-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho D.S., Yim H., Cho K.S., Hong S.J., Cho N.H., Kim S.I., Ahn H.S., Kim S.J. Impact of caveolin-1 expression on the prognosis of transitional cell carcinoma of the upper urinary tract. J Korean Med Sci. 2008;23:296–301. doi: 10.3346/jkms.2008.23.2.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kunze E., Von Bonin F., Werner C., Wendt M., Schlott T. Transitional cell carcinomas and nonurothelial carcinomas of the urinary bladder differ in the promoter methylation status of the caveolin-1, hDAB2IP and p53 genes, but not in the global methylation of Alu elements. Int J Mol Med. 2006;17:3–13. [PubMed] [Google Scholar]
- 57.Hanke M., Kausch I., Dahmen G., Jocham D., Warnecke J.M. Detailed technical analysis of urine RNA-based tumor diagnostics reveals ETS2/urokinase plasminogen activator to be a novel marker for bladder cancer. Clin Chem. 2007;53:2070–2077. doi: 10.1373/clinchem.2007.091363. [DOI] [PubMed] [Google Scholar]
- 58.Lee S.J., Park S.S., Cho Y.H., Park K., Kim E.J., Jung K.H., Kim S.K., Kim W.J., Moon S.K. Activation of matrix metalloproteinase-9 by TNF-alpha in human urinary bladder cancer HT1376 cells: the role of MAP kinase signaling pathways. Oncol Rep. 2008;19:1007–1013. [PubMed] [Google Scholar]
- 59.Morrisey E.E., Ip H.S., Tang Z., Lu M.M., Parmacek M.S. GATA-5: a transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol. 1997;183:21–36. doi: 10.1006/dbio.1996.8485. [DOI] [PubMed] [Google Scholar]
- 60.Cohen A.W., Park D.S., Woodman S.E., Williams T.M., Chandra M., Shirani J., Pereira de Souza A., Kitsis R.N., Russell R.G., Weiss L.M., Tang B., Jelicks L.A., Factor S.M., Shtutin V., Tanowitz H.B., Lisanti M.P. Caveolin-1 null mice develop cardiac hypertrophy with hyperactivation of p42/44 MAP kinase in cardiac fibroblasts. Am J Physiol Cell Physiol. 2003;284:C457–C474. doi: 10.1152/ajpcell.00380.2002. [DOI] [PubMed] [Google Scholar]
- 61.Williams T.M., Medina F., Badano I., Hazan R.B., Hutchinson J., Muller W.J., Chopra N.G., Scherer P.E., Pestell R.G., Lisanti M.P. Caveolin-1 gene disruption promotes mammary tumorigenesis and dramatically enhances lung metastasis in vivo: role of Cav-1 in cell invasiveness and matrix metalloproteinase (MMP-2/9) secretion. J Biol Chem. 2004;279:51630–51646. doi: 10.1074/jbc.M409214200. [DOI] [PubMed] [Google Scholar]
- 62.Williams T.M., Lisanti M.P. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol. 2005;288:C494–C506. doi: 10.1152/ajpcell.00458.2004. [DOI] [PubMed] [Google Scholar]
- 63.Bergdahl A., Swärd K. Caveolae-associated signaling in smooth muscle. Can J Physiol Pharmacol. 2004;82:289–299. doi: 10.1139/y04-033. [DOI] [PubMed] [Google Scholar]
- 64.Lee Y.H., Hwang M.K., Morgan K.G., Taggart M.J. Receptor-coupled contractility of uterine smooth muscle: from membrane to myofilaments. Exp Physiol. 2001;86:283–288. doi: 10.1113/eph8602184. [DOI] [PubMed] [Google Scholar]


