Abstract
RSF-1, also known as hepatitis B X-antigen associated protein (HBXAP), is a subunit of an ISWI chromatin remodeling complex, remodeling and spacing factor (RSF). Recent studies have provided new evidence that chromatin remodeling participates in the pathogenesis of neoplastic diseases by altering cell cycle regulation and gene expression. In this report, we studied the biological roles of RSF-1 in oral squamous cell carcinoma (OSCC), a highly invasive neoplastic disease. Based on IHC and quantitative real-time PCR, we demonstrated that RSF-1 expression could be detected in the majority of OSCC cases, and the levels were significantly higher in OSCC cells than in their normal counterparts. Moreover, expression levels of RSF-1 significantly correlated with the presence of angiolymphatic invasion, abnormal mitoses, metastasis, tumor recurrence, and advanced stage disease at presentation. Univariate and multivariate analyses showed a significant association of RSF-1 overexpression and worse overall survival in OSCC patients. RSF-1 knockdown remarkably decreased cellular proliferation and induced apoptosis in OSCC cells with high RSF-1 expression levels, but not in those without. Taken together, our results suggest that RSF-1 up-regulation is associated with several clinicopathological features of disease aggressiveness in OSCC patients, and RSF-1 plays an important role in maintaining cellular growth and survival in OSCC.
Genomic DNA in eukaryotic nuclei is highly organized and packaged with histone proteins into the basic building blocks of chromatin (ie, nucleosomes). Chromatin structure is highly dynamic, allowing DNA synthesis, regulation of transcription, and damage repair in a highly efficient fashion. Such processes are made possible by chromatin remodeling factors that are essential for packaging and unwinding regions of the chromosome in response to a variety of cellular signaling and biological cues, allowing access by specific nuclear proteins.1 Chromatin remodeling plays a fundamental role in cellular biology, including tissue development and differentiation, as well as contributing to the pathogenesis of a variety of diseases including cancer.2–4 There are at least three main families of chromatin remodeling complexes including the ISWI, SWI/SNF, and CHD/Mi-2 families.4 Inappropriate chromatin remodeling activity can lead to dysregulated gene activation that is associated with oncogenesis.
Several chromatin remodeling genes have been reported to be involved in human cancer. For example, somatic mutations of ARID1A (BAF250A) have been recently demonstrated in approximately 50% of ovarian clear cell carcinomas.5,6 Besides, potential tumor suppressor roles have been demonstrated for ATPase-containing factors, including Brg and ARID1A, which are components of the SWI/SNF chromatin remodeling complex.7–9 On the other hand, a tumor-promoting function has been reported for RSF1, also known as hepatitis B X-antigen associated protein (HBXAP), which is a histone binding protein that interacts with sucrose nonfermenting protein 2 homologue (SNF2H) to form a complex belonging to the ISWI chromatin remodeling family.10–12 We have demonstrated that RSF1 is located in the ch11q13.5 region, which is frequently amplified in several types of carcinoma, including ovarian high-grade serous carcinoma.10,13,14 As compared to other genes within the ch11q13.5 amplicon, RSF1 exhibited the highest correlation between DNA amplification and RNA copy number, and it was responsible for cell growth in the presence of chemotherapeutic agents in ovarian cancer tissues,10,15 suggesting that RSF1 serves as the “driver” gene within the ch11q13.5 amplicon. RSF-1 overexpression was highly associated with the most aggressive type of ovarian cancer, high-grade serous carcinoma, and shorter overall survival.10,16,17
In the current report, we focused on determining the biological significance of RSF-1 expression in oral squamous cell carcinoma (OSCC), the most common oral malignancy. Based on IHC, using a well-characterized antibody, we correlated RSF-1 expression levels with clinicopathological features associated with aggressiveness of the disease. In addition, to assess the efficacy of targeting RSF-1 as a potential therapeutic strategy, we studied the effect of RSF-1 knockdown in OSCC cell lines. These studies demonstrated an essential role of RSF-1 in maintaining cellular proliferation and survival in RSF-1-overexpressing OSCC cells.
Materials and Methods
Tissue Collection and TMAs
Tumor tissues were obtained from the Chang Gung Memorial Hospital-Kaohsiung Medical Center and the China Medical University Hospital in Taiwan. Tissue specimens and clinical information were acquired in accordance with the regulations of the institutional review board. A group of 98 OSCC tissue specimens, 18 tumor-adjacent epithelium samples, including 8 with carcinoma in situ (CIS) and 10 without significant dysplasia, as well as 10 epithelial samples collected from healthy individuals without OSCC history were enrolled for RSF-1 immunostaining. An independent cohort that included 15 pairs of primary and recurrent OSCCs from individual patients were analyzed. To facilitate IHC, 1.5-mm cores from each specimen were arranged in triplicate in TMAs.
Immunohistochemistry
The TMA slides were subjected to antigen retrieval by boiling the slides in citrate buffer, pH 6.0 (Zymed, South San Francisco, CA) for 20 minutes. Samples were then stained with anti-RSF-1 antibodies (Upstate, Lake Placid, NY) at a 1:1000 dilution at room temperature for 1 hour. An EnVision+ Systems Peroxidase kit (DAKO, Carpentaria, CA) was used for chromogen development. The specificity of anti-RSF-1 antibody for use in IHC has been previously demonstrated.10,17,18 A positive reaction was defined as discrete localization of the brown chromogen in the nuclei. Immunostaining intensity was independently scored by two investigators using a previously described scoring system10; intensity was recorded as negative (0), weakly positive (1+), moderately positive (2+), strongly positive (3+), or intensely positive (4+). In discordant cases, the sample was scored by a third investigator and the final score was determined by the majority scores.
DNA/RNA Extractions and Quantitative Real-Time PCR
Tumor cell content of samples was determined from histological examination of frozen sections. Specimens were selected for DNA/RNA extractions and quantitative real-time PCR only when the tissues contained at least 85% tumor cells. DNA and RNA were purified from the selected samples using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the procedure provided by the manufacturer. The RNA samples were reverse-transcribed into cDNA using MMLV RT enzymes (Promega, Madison, WI). Primers for quantitative PCR for RSF1 expression were designed using the primer 3 program (http://frodo.wi.mit.edu/primer3, last accessed March 8, 2011). The primer sequences for RSF-1 were: forward, 5′-GAGGAGGATGCCGATACTATGC-3′; and reverse, 5′-TGCTTTCAGGAGTGCAAGAGTC-3′. Quantitative PCR was performed on cDNA samples from 48 independent OSCC tissues and 10 normal oral epithelia. PCR reactions were performed in duplicate and threshold cycle numbers were calculated using iCycler software v2.3 (Bio-Rad Laboratories, Hercules, CA). The expression level of RSF-1 for each sample was determined by normalization to the expression level of glyceraldehyde-3-phosphate dehydrogenase. The primer sequences for glyceraldehyde-3-phosphate dehydrogenase were: forward, 5′-AGCCACATCGCTCAGACAC-3′; and reverse, 5′-GCCCAATACGACCAAATCC-3′. To detect the DNA copy number of RSF1 gene, quantitative PCR was performed on genomic DNA samples from 30 independent OSCC tissues and 8 normal oral epithelia. The primer sequences were: forward, 5′-TCTTTAAGCATGGCTGTCCAAA-3′; and reverse, 5′-AGCCATGGAAGGAAATAGCTGA-3′. DNA content of RSF1 gene in each sample was then normalized to that of LINE-1, as previously described.19
Statistical Analysis and Clinical Correlations
Clinicopathological information, including angiolymphatic invasion, metastasis, clinical stage, treatment history, recurrent status, and survival data, was recorded and correlated with RSF-1 expression levels (Table 1). Overall survival time was defined as the number of months between the primary diagnosis and death or between diagnosis and the most recent follow-up. Survival was adjusted for clinical stages, age (median, 68 years; range, 34 to 87 years) and tumor location. All calculations and statistical analyses were performed using SPSS 14.0 (SPSS Inc., Chicago, IL) software, and the association and comparison between various clinicopathological factors and RSF-1 expression were assessed by χ2 test, Mann-Whitney U-test, Kruskal-Wallis H test, and paired t-test, as appropriate. Survival curves were plotted using the Kaplan-Meier method. With this method, P values were evaluated by the log-rank test. All significant parameters (P < 0.05) at the univariate level were entered into a Cox regression model to analyze their relative prognostic importance. For all analyses, two-sided tests of significance were used, with P < 0.05 considered significant.
Table 1.
Summary of Clinicopathological Features of 98 Patients
| Parameters | Number of cases (%) |
|---|---|
| Sex | |
| Male | 97 (99.0%) |
| Female | 1 (1.0%) |
| Age (years) (Median, 68; range, 34–87) | |
| ≤40 | 20 (20.4%) |
| 41–50 | 35 (35.7%) |
| 51–60 | 30 (30.6%) |
| 61–70 | 10 (10.2%) |
| 71–80 | 3 (3.1%) |
| 81–90 | 1 (1.0%) |
| Tumor location | |
| Tongue | 40 (40.8%) |
| Buccal | 37 (37.8%) |
| Others | Lip (6), mouth floor (6), palate (5), trigone (4) |
| Primary tumor (T) | |
| T1 | 18 (18.4%) |
| T2 | 31 (31.6%) |
| T3 | 17 (17.3%) |
| T4 | 32 (32.7%) |
| Nodal status (N) | |
| N0 | 38 (46.3%) |
| N1 | 14 (17.1%) |
| N2 | 30 (36.6%) |
| Nx | 16 |
| Extracapsular extension of metastatic nodes | |
| Present | 32 (61.5%) |
| Absent | 20 (38.5%) |
| Histological grade | |
| Well differentiated | 61 (62.2%) |
| Moderately differentiated | 29 (28.6%) |
| Poorly differentiated | 9 (9.2%) |
| Vascular invasion | |
| Present | 18 (18.4%) |
| Absent | 80 (81.6%) |
| Perineurial invasion | |
| Present | 26 (26.5%) |
| Absent | 72 (73.5%) |
| Tumor necrosis | |
| Present | 38 (38.8%) |
| Absent | 60 (61.2%) |
| CIS at adjacent mucosa | |
| Present | 24 (24.5%) |
| Absent | 74 (75.5%) |
| Surgical margin | |
| Clear (≥5 mm) | 91 (94.8%) |
| Unclear | 5 (5.2%) |
| Not assessable | 2 |
| RSF-1 expression | |
| Low expression (0+∼2+) | 44 (44.9%) |
| Overexpression (3+∼4+) | 54 (55.1%) |
CIS, carcinoma in situ.
siRNA-Mediated Knockdown Assay
Three short-interfering RNA (siRNAs) that targeted RSF-1 were designed; their sense sequences were: 5′-GGAAAGACAUCUCUACUAUUU-3′, 5′-UAAAUGAUCUGGACAGUGAUU-3′, and 5′-GGACUUACCUUCAACCAAUUU-3′. Scramble siRNA (off-target control, catalog no. D-001210-02-05) was purchased from Dharmacon (Lafayette, CO). Cells were seeded in 96 wells and transfected with a mixture of the three RSF-1 siRNAs using oligofectamine (Invitrogen). Growth and phenotypes of treated cells were monitored for 4 days.
Western Blot Analysis
Protein lysates were prepared from oral cancer cell lines, including KON, SAT, SSC4, SSC25, HSC3, and CAL27 cells. KON and SAT cells were purchased from the Japanese Collection of Research Bioresources (JCRB, Shinjuku, Japan). SSC4, SSC25, HSC3, and CAL27 cells were purchased from the Bioresource Collection and Research Center (BCRC, Taiwan). Proteins were separated by SDS-PAGE and transferred onto PVDF membranes. Western blots were performed by hybridizing the membranes with an anti-RSF-1 antibody and an anti-glyceraldehyde-3-phosphate dehydrogenase antibody for 2 hours at room temperature, at a dilution of 1:1000. After three washes with TBST (0.01% Tween 20 in TBS), the membranes were blotted with HRP-conjugated anti-mouse antibodies (Pierce, Rockford, IL), at a dilution of 1:1000 for 1 hour at room temperature. RSF-1 bands were revealed by chemiluminescence (Amersham Biosciences, Arlington Heights, IL).
Cell Proliferation Activity and Apoptosis Assay
To determine the effects of RSF-1 knockdown, cell number was determined indirectly by fluorescence intensity of SYBR Green I nucleic acid stain (Molecular Probes, Eugene, OR), as detected with a microplate reader (Fluostar, BMG, Durham, NC). Data were expressed as mean ± SD from five replicates in each experimental group. To study DNA synthesis rate, bromodeoxyuridine (BrdU) uptake and staining were performed using a cell proliferation kit (Amersham). Briefly, cells were incubated with 10 μmol/L BrdU at 37°C for 4 hours and collected by trypsinization. After fixation with 70% ethanol, cells were spotted on slides using a cytospin (Thermo, Pittsburgh, PA). The slides were stained with the anti-BrdU antibody and counterstained with DAPI. Apoptotic cells were detected by staining with annexin V-FITC (BioVision, Mountain View, CA) 4 days after transfection. The percentages of BrdU-positive and annexin V-positive cells were determined by counting at least 400 randomly selected cells for each experiment.
Results
Up-Regulation of RSF-1 Is Common in Oral Squamous Cell Carcinoma
IHC was performed in 98 primary OSCC tissues to correlate RSF-1 expression levels with clinicopathological features. Consistent with our previous studies on other cancer types,10,16,17 RSF-1 immunoreactivity was exclusively detected in the nuclei (Figure 1A). RSF-1 immunoreactivity was detected in the majority of OSCC samples, and was undetected in only 8 (8.2%) of 98 OSCC samples. In contrast, a staining pattern from undetectable to a weak RSF-1 staining was found in oral surface squamous epithelium from normal controls (P < 0.001) compared with OSCCs (see Supplemental Figure S1A at http://ajp.amjpathol.org). Of note, carcinoma in situ (CIS) tissues demonstrated a significantly higher RSF-1 expression level than that in both normal controls and non-neoplastic tumor-adjacent epithelia (P = 0.013 and P = 0.020, respectively) (see Supplemental Figure S1B at http://ajp.amjpathol.org). RSF-1 levels did not show difference between non-neoplastic tumor-adjacent epithelium and normal control (P = 0.375). Among the samples tested, 54 of 98 (55.1%) OSCC samples were scored as +3 or +4 for RSF-1 staining, indicating high expression, whereas 36 of 98 (36.7%) tumor samples received immunostaining scores of +1 or +2 indicating low RSF-1 expression. To further assess the clinical significance of RSF-1 expression in OSCC, we validated the IHC data by reverse transcription-quantitative PCR. Our results indicated a higher average level (∼2.7-fold) of RSF-1 expression in OSCC than in normal squamous epithelial tissues (P = 0.006) (Figure 1B). In the present study, RSF gene was also found amplified in OSCC (P = 0.042) (Figure 1C). Similar to the findings in ovarian cancer,10 OSCC samples with higher RSF-1 DNA copy number tended to show higher expression levels, suggesting that RSF-1 was frequently amplified and overexpressed in OSCC tissues at both the protein and mRNA levels.
Figure 1.
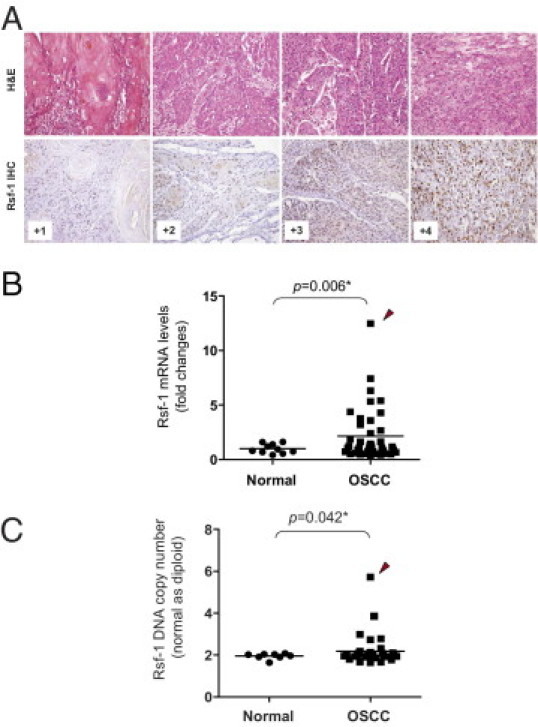
RSF1 gene is overexpressed in oral squamous cell carcinoma (OSCC). A: Representative examples of OSCC were stained with H&E. Based on IHC, RSF-1 immunoreactivity on tumor tissues was scored as 0, +1, +2, +3, or + 4. B: Quantitative real-time PCR (qPCR) analysis indicated higher expression levels of the RSF1 gene in OSCC than in normal epithelial cells. C:RSF1 amplification was frequently found in OSCC tissues analyzed by qPCR. Arrowhead indicates the OSCC sample with highest RSF1 content and gene expression level.
RSF-1 Expression Level Correlates with Poor Clinical Outcome
Our previous studies showed that overexpression of RSF-1 correlated with poor clinical outcomes in ovarian cancer patients.10,16,17 Based on the criteria for immunostaining scoring and sample classification in these studies (see Materials and Methods), we stratified cases into two groups: i) with low RSF-1 expression (scores of 0 to +2), and ii) with high RSF-1 expression (scores of +3 or +4). Kaplan-Meier survival analysis was performed to assess the association of RSF-1 expression with different outcomes in overall patient survival. Univariate and multivariate analyses showed that RSF-1 overexpression (P = 0.001), lymph node metastasis (P = 0.012), and clinical stage of primary tumors (P = 0.044) were associated with shorter disease-associated survival times (Table 2). As shown in Figure 2A, patients with high-expression levels of RSF-1 had a shorter overall survival time than did patients with low levels of RSF-1, suggesting that RSF-1 overexpression is an indicator of poor prognosis for OSCC patients. Furthermore, we found that the RSF-1 overexpression was significantly associated with an increment of the status of pathological classification (pT status) with a P value of 0.025 (Figure 2B). These findings confirmed that RSF-1 overexpression was associated with more aggressive OSCC.
Table 2.
Univariate and Multivariate Analyses for Disease-Specific Survival
| Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| Parameters | Number of cases | Number of events | P value | HR | 95% CI | P value |
| Sex | ||||||
| Male | 97 | 40 | 0.4239 | |||
| Female | 1 | 0 | ||||
| Age (years) | ||||||
| <60 | 85 | 37 | 0.2537 | |||
| ≥60 | 13 | 3 | ||||
| Primary tumor (T) | ||||||
| T1-T2 | 49 | 10 | 0.0008⁎ | 1 | 0.044⁎ | |
| T3 | 17 | 10 | 1.303 | 0.562–3.021 | ||
| T4 | 32 | 20 | 2.439 | 1.009–5.882 | ||
| Nodal status (N) | ||||||
| N0 | 38 | 9 | 0.0009⁎ | 1 | 0.012⁎ | |
| N1 | 14 | 8 | 1.488 | 0.614–63.610 | ||
| N2 | 30 | 19 | 2.849 | 1.258–6.452 | ||
| Extracapsular extension of metastatic node | ||||||
| Present | 20 | 12 | 0.2890 | |||
| Absent | 32 | 18 | ||||
| Histological grade | ||||||
| Well differentiated | 61 | 22 | 0.2558 | |||
| Moderately differentiated | 28 | 13 | ||||
| Poorly differentiated | 9 | 5 | ||||
| Vascular invasion | ||||||
| Present | 18 | 10 | 0.1461 | |||
| Absent | 80 | 30 | ||||
| Perineurial invasion | ||||||
| Present | 26 | 14 | 0.1553 | |||
| Absent | 72 | 26 | ||||
| Tumor necrosis | ||||||
| Present | 38 | 17 | 0.5545 | |||
| Absent | 60 | 23 | ||||
| CIS at adjacent mucosa | ||||||
| Present | 24 | 12 | 0.2476 | |||
| Absent | 74 | 28 | ||||
| Surgical margin | ||||||
| Clear | 91 | 37 | 0.9199 | |||
| Unclear | 5 | 2 | ||||
| RSF-1 expression | ||||||
| Low expression | 44 | 3 | <0.0001⁎ | 1 | 0.001⁎ | |
| Overexpression | 54 | 37 | 33.97 | 4.456–244.598 | ||
Follow-up duration ranged from 1 to 112 months; mean, 40.12; median, 45.
95% CI, 95% confidence interval; CIS, carcinoma in situ; HR, hazard ratio.
Statistically significant.
Figure 2.
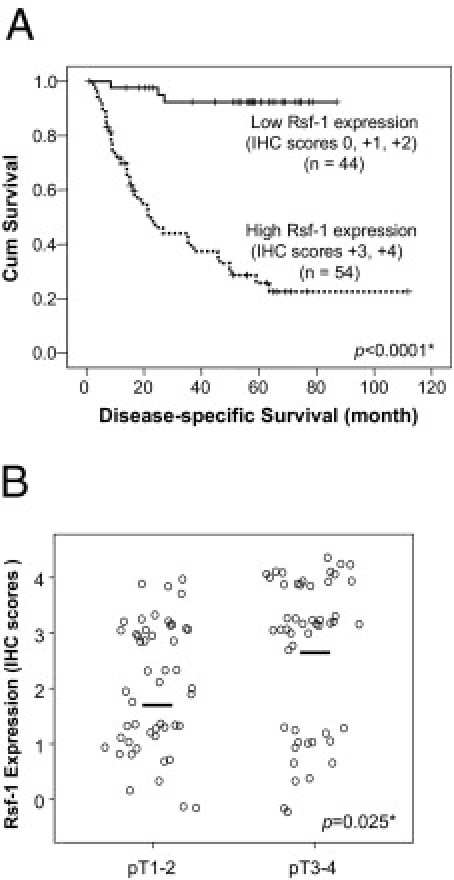
RSF-1 expression correlates with poor clinical outcome in oral squamous cell carcinoma (OSCC). A: Kaplan-Meier survival analysis shows an association between RSF-1 protein expression and shorter overall survival. Patients with immunostaining scores of 0 to + 2 were considered to have low RSF-1 expression; patients with scores of +3 or +4 were considered to have high RSF-1 expression. B: Correlation analysis of RSF-1 expression level and pathological pT status demonstrates that cases with higher pT stages (pT3 and pT4) have higher RSF-1 levels than do lower stage cases (pT1 and pT2).
Involvement of RSF-1 Expression in Advanced Cancer Development
Because OSCC is a highly invasive disease that displays frequent tumor metastasis and recurrence, we asked if there was a link between RSF-1 expression levels in OSCC tissues and aggressive tumor behaviors. Microscopically, tumors that highly expressed RSF-1 were more frequently associated with deeper invasion, with a significant increment in tumor thickness than those with RSF-1 low expression (17.02 ± 12.1 mm and 11.61 ± 10.9 mm, respectively; P = 0.011). Tumors demonstrated an adverse invasion pattern, which was defined by invasive tumor front classification (ITFc) grades 3 and 4 and had increased RSF-1 immunostaining intensity than those with low grade (P = 0.027). In fact, increased RSF-1 immunostaining intensity in OSCC samples was significantly associated with the presence of angiolymphatic tumor invasion (P = 0.001) (Figure 3A), and lymph node metastasis (P = 0.027) (Figure 3B). Moreover, RSF-1 overexpression was also significantly associated with the presence of abnormal mitotic figures including tri-polar or tetra-polar metaphase and anaphase bridges (P = 0.033) (Figure 3C). To assess RSF-1 involvement in the development of recurrent tumors, we compared RSF-1 immunostaining intensities between primary and recurrent OSCC samples from 15 cases for which paired samples were available (Figure 4A). Our results showed that recurrent tumors had higher RSF-1 expression levels than the paired primary tumors (P = 0.001) (Figures 4, A and B). These findings suggested that RSF-1 overexpression was associated with tumor progression.
Figure 3.
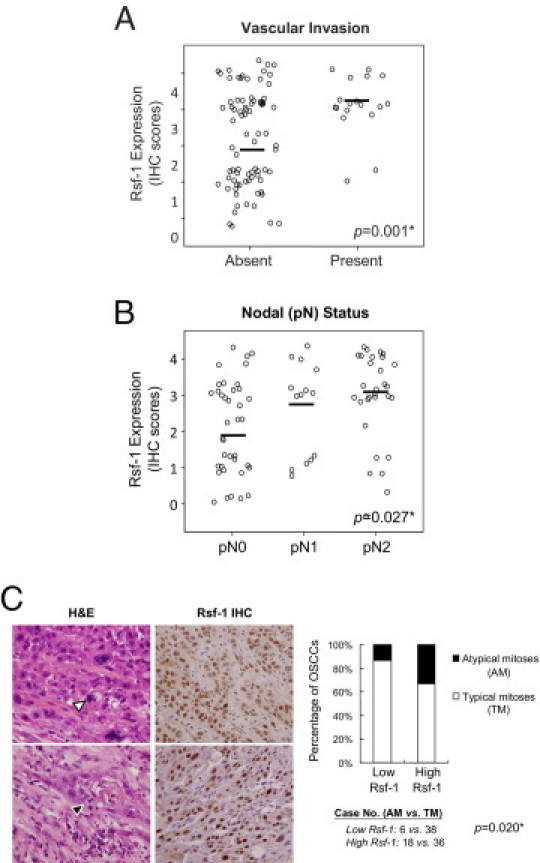
RSF-1 expression correlates with more tumor aggressiveness in oral squamous cell carcinoma (OSCC). Higher expression levels of RSF-1 were significantly associated with vascular invasion (A) and metastasis (B) to the regional lymph nodes. C: High RSF-1 expressing OSCC samples (scores +3 or +4) showed more frequent atypical mitoses than OSCC samples with low RSF-1 expression (scores 0, +1, or +2). Two pairs of representative H&E and RSF-1 staining tissue sections showed an abnormal number of spindle pole (white arrowhead) and anaphase bridge (black arrowhead), respectively. H&E (hematoxylin and eosion) section with magnification of 40× and RSF-1 IHC (immunohistochemistry) section of 10×.
Figure 4.
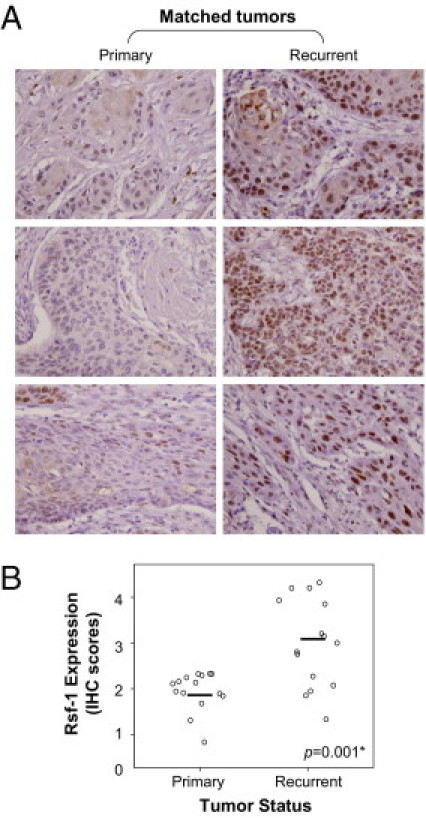
RSF-1 overexpression is associated with recurrent disease. A: Representative paired primary and recurrent oral squamous cell carcinoma (OSCC) tissues are shown. Stronger nuclear RSF-1 staining was observed in recurrent OSCC. Magnification: ×20 B: Immunohistochemical analyses of 15 paired OSCC tumors from the same patients.
The Essential Role of RSF-1 in Cellular Proliferation and Survival in OSCC Cells with RSF-1 Overexpression
To assess the possibility that RSF-1 overexpression was associated with molecular dependence of the tumor on RSF-1 for cellular growth and survival, we compared the phenotypes of OSCC cells (with high basal RSF-1 expression) before and after RSF-1 knockdown with the phenotypes of cells with low or undetectable basal RSF-1 expression. First, protein levels of RSF-1 in 6 OSCC cell lines were analyzed using Western blotting (Figure 5A). Among the cell lines tested, SAT cells were found to contain the most abundant RSF-1 protein followed by KON cells, whereas SSC4 and CAL27 cells showed very weak or undetectable RSF-1 levels. SSC25 and HSC3 cells demonstrated intermediate levels of RSF-1 expression. We then performed RSF-1 knockdown with short-interfering RNA (siRNA) against RSF-1 and compared cell growth and apoptotic activity in four cell lines: SAT, KON, SSC4, and CAL27. As shown in Figure 5B, RSF-1 protein levels could be substantially reduced by RSF-1 siRNA in SAT cells, indicating the robust effect of the siRNA knockdown. Cell growth analyses revealed a significant decrease in numbers of SAT cells at all time points (P = 0.0032) after siRNA transfection, whereas there was only a moderate effect on KON cells and minimal effects on SSC4 and CAL27 cells (a pattern corresponding to their RSF-1 protein levels) (Figure 5C). These results suggested that basal RSF-1 expression levels in oral cancer cells determined cellular sensitivity to RSF-1 knockdown.
Figure 5.
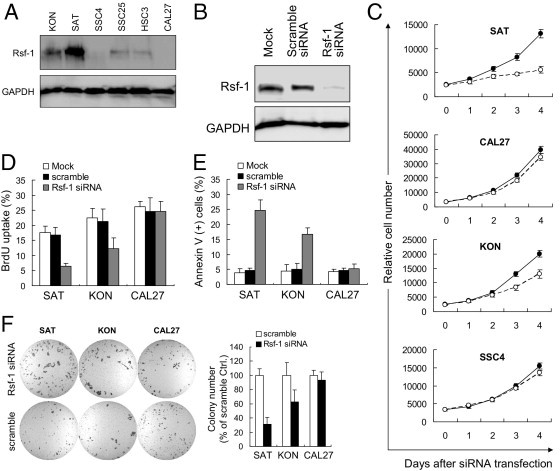
Assessment of RSF1 gene knockdown in RSF-1 expressing oral squamous cell carcinoma (OSCC) cells. A: Western blot analysis revealed the highest expression level of RSF-1 in SAT cells, a moderate level in KON cells, and undetectable levels in SSC4 and CAL27 cells. B: Western blot analysis shows an efficient reduction of RSF-1 protein by RSF-1 specific short-interfering RNA (siRNA). Cells treated with buffer alone (mock) or scramble siRNA were used as the controls. C: Differential growth suppression was found when cells were treated with RSF-1 specific siRNA (blank circles). Cells treated with scramble siRNA were used as the controls (filled circles). DNA synthesis was monitored by (D) Bromodeoxyuridine (BrdU) incorporation and (E) cell death induction was monitored by annexin V staining 4 days after RSF-1-specific siRNA treatment. Cells treated with buffer alone (mock) or scramble siRNA were used as controls. F: Soft agar assay indicates that RSF-1 knockdown leaded to reduction of colony formation in SAT and KON cells, but caused very minor effects on CAL27 cells. Cells treated with scramble served as controls.
The growth inhibitory effect of RSF-1 siRNA in SAT and KON cells was found to be largely due to the inhibition of DNA synthesis, because the percentage of BrdU-labeled cells was significantly decreased in RSF-1 siRNA-treated SAT and KON cells as compared to mock treated cells (P < 0.0001 and P = 0.003, respectively) or scramble siRNA treated cells (P = 0.0001 and P = 0.002, respectively) (Figure 5D). In addition, inhibition of cellular growth by RSF-1 knockdown was followed by apoptosis in RSF-1 siRNA-treated SAT and KON cells, which were measured by annexin V staining (Figure 5E). Increased apoptotic activity was only observed 4 days after transfection. Control cells treated with mock or scramble siRNA showed minimal cell death. RSF-1 knockdown had minimal effect on growth or apoptosis of CAL27 cells under the same experimental conditions. Soft agar assays further indicated survival advantages of RSF-1 signaling in RSF-1-overexpressing OSCC cells, as RSF-1 knockdown significantly reduced anchorage-independent growth in SAT and KON cells, but not in CAL27 cells. These findings suggested a potent role for RSF-1 in OSCC pathogenesis, and demonstrated that RSF-1 knockdown significantly blocked tumor-promoting activity in RSF-1 expressing OSCC cells, and resulted in growth arrest and subsequent cell death.
Discussion
Oral cancer is the third most common cancer in developing countries and the sixth most common cancer worldwide.20,21 Among different types of oral cancer, OSCC represents one of the most common types, and are frequently associated with repeated mucosal damage and sustained inflammation. Although oral cancer can be cured when the tumor is detected at an early stage, patients who have had oral cancers have a high risk of developing secondary and/or recurrent tumors in the surrounding oral mucosa, presenting a major challenge in the management of OSCC patients. Once tumor cells spread to lymph nodes, the overall mortality rate is high, and has not been significantly improved in decades. A better understanding of the molecular pathways involved in oral tumorigenesis could provide a roadmap toward improving the clinical outcome of OSCC. The pathogenesis of OSCC remains poorly understood, but recent studies have provided new clues into how it develops. For example, using a genome-wide analysis of DNA copy number changes, we have previously demonstrated that amplification at the ch7p11.2 locus, which harbors epidermal growth factor receptor (EGFR), was the most common DNA copy number alteration in OSCC tissues.19 Up-regulation of other known oncogenes, such as CCND1 and FGFR genes, was also frequently found in OSCC.19,22,23 In this study, we demonstrate that a chromatin remodeling factor, RSF-1, may be involved in tumor progression, and may confer disease aggressiveness in OSCC. Moreover, RSF-1 knockdown experiments showed that RSF-1 expression was essential for tumor cell growth and survival in RSF-1-overexpressing OSCC cells.
The results from this report have several biological implications that may shed new light on the molecular etiology of OSCC. Similar to other cancer types, such as ovarian high-grade serous carcinoma10,16,17 and ovarian clear cell carcinoma,18 up-regulation of RSF-1 is associated with the aggressiveness of OSCC, including tumor invasion, lymph node metastasis, and rapidly advancing clinical stage. Multivariate analysis has shown that RSF-1 overexpression is an independent marker for a worse overall survival in OSCC patients. Currently, overexpression of transforming growth factor-α and EGFR in OSCC has been reported to have a strong prognostic value,19,24 suggesting that RSF-1 immunoreactivity may serve as another OSCC biomarker for clinical outcome prediction. It would be of great interest to determine whether analysis of expression of the combination of RSF-1, transforming growth factor-α, and EGFR would further improve outcome prediction. Classifying patients into high-risk and low-risk groups for aggressive disease could help when making treatment decisions in the management of OSCC patients. The high-risk group may warrant close monitoring for secondary and recurrent tumors. Those patients may also be considered for a more aggressive treatment to prevent disease progression.
Although more efforts are needed to determine exactly how RSF-1 proteins contribute to the aggressive phenotypes in OSCC tissues, one possibility (suggested by the observation that recurrent OSCC showed higher RSF-1 levels than did matched primary tumors) is that RSF-1 potentiates resistance to radiotherapy or chemotherapy. This view is supported by our previous study showing that RSF-1 can enhance drug resistance through remodeling and spacing factor complex formation in ovarian cancer cells.15 Alternatively, RSF-1 overexpression may directly contribute to tumor invasiveness and metastasis. In fact, recent studies have indicated that certain proteins contributing to metastasis, such as metastasis-associated gene in NuRD complex25 or MYC and BRCA1 in SWI/SNF complex,26,27 can form functional complexes with chromatin remodeling factors in promoting cancer progression.
At the molecular level, it has been demonstrated that regulation of nucleosome deposition is essential for transcriptional activation or repression,28–30 DNA replication,31,32 DNA damage repair,4,33,34 and cell cycle progression.35–37 Chromatin remodeling factors that control DNA winding/unwinding cycles are therefore believed to be involved in these processes. Recent reports have even proposed ATP-dependent chromatin remodeling factors as one category of cell cycle enzymes.36 In addition to RSF-1, a number of genes involved in chromatin remodeling were also reported to be associated with cancer development. For example, chromosomal rearrangements of chromatin regulators SMARCC1, SMARCC2, and SMARCD1 were frequently found in human lung cancer and in a variety of solid tumors.38,39 A novel alternative splicing variant of the SNF2-like gene PASG was found in >25% of leukemia and lymphoma patients.40 In our recent study, overexpression of RSF-1 frequently caused DNA double-strand breaks in nontransformed normal cells, which subsequently lead to activation of DNA damage responses and severe cell death.41 However, RSF-1-induced DNA damage responses can serve as a selection barrier for cell clones with genetic defects in the genes involved in DNA damage responses, thus allow further genetic alterations and oncogene activation during cancer development. Our results in the current study provide evidence that dysregulation of chromatin structure by up-regulation of RSF-1 expression could be one of the mechanisms to promote oral cancer development.
In conclusion, our data suggest that chromatin remodeling factor, RSF-1, participates in the tumor progression of OSCC and could be considered as a prognostic marker for predicting clinical outcome. Our data also showed that RSF-1 overexpression is associated with tumor invasion and recurrence, which is the main cause of high mortality among patients with advanced oral cancers. As with ovarian high-grade serous carcinoma,10,11 RSF-1 knockdown leads to growth suppression and cell death in those tumors cells with a high level of RSF-1 up-regulation. The molecular dependence on RSF-1 protein for maintaining cellular proliferation and survival is of potential translational interest. Targeting RSF1 gene expression and the pathway it controls may provide new therapeutic avenues for treating advanced stage OSCC that are refractory to conventional therapy.
Acknowledgments
The authors thank the thoughtful suggestions and comments on this article from Dr. Lei Wan and Dr. Ying-Ju Lin (China Medical University, Taichung, Taiwan). We also thank Ms. Yi-Ting Hsiao and Ms. Carmen Chan (China Medical University Hospital, Taichung, Taiwan) for their technical assistance.
Footnotes
This study was supported in part by funds from the National Institutes of Health/National Cancer Institute grant RO1CA129080 (I.M.S.), the National Science Council, Taiwan grant NSC96-2321-B-182A-001-MY2 (C.F.L.) and grant NSC98-2320-B-039-033-MY3 (J.J.S.), the Department of Health, Taiwan grant DOH99-TD-C-111-004 (J.J.S.), and the China Medical University, Taiwan grant CMU97-280 (J.J.S.).
F.M. Fang and C.F. Li equally contributed to this article.
Supplemental material for this article can be found at http://ajp.amjapthol.org or at doi: 10.1016/j.ajpath.2011.01.043.
Contributor Information
Ie-Ming Shih, Email: ishih@jhmi.edu.
Jim Jinn-Chyuan Sheu, Email: jimsheu@mail.cmu.edu.tw.
Supplementary data
Immunoreactivity of anti-Rsf-1 in oral squamous cell carcinoma tissues and cells. A: Immunoreactivity was weak or undetectable in normal control and non-neoplastic tumor-adjacent epithelial cells. B: A stepwise and significant increment of Rsf-1 expression in non-neoplastic epithelium, CIS, and tumor cells could also be discerned.
References
- 1.Workman J.L., Kingston R.E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Wolffe A.P. Chromatin remodeling: why it is important in cancer. Oncogene. 2001;20:2988–2990. doi: 10.1038/sj.onc.1204322. [DOI] [PubMed] [Google Scholar]
- 3.Lafon-Hughes L., Di Tomaso M.V., Mendez-Acuna L., Martinez-Lopez W. Chromatin-remodelling mechanisms in cancer. Mutat Res. 2008;658:191–214. doi: 10.1016/j.mrrev.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Wang G.G., Allis C.D., Chi P. Chromatin remodeling and cancer. Part II: aTP-dependent chromatin remodeling. Trends Mol Med. 2007;13:373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiegand K.C., Shah S.P., Al-Agha O.M., Zhao Y., Tse K., Zeng T., Senz J., McConechy M., Anglesio M.S., Kalloger S.E., Yang W., Heravi-Moussavi A., Giuliany R., Chow C., Fee J., Zayed A., Melnyk N., Turashvili G., Delaney A., Madore J., Yip S., McPherson A.W., Ha G., Bell L., Fereday S., Tam A., Galletta L., Tonin P.N., Provencher D., Miller D., Jones S., Moore R.A., Morin G.B., Oloumi A., Boyd N., Aparicio S.A., Shih I.M., Mes-Masson A., Bowtell D., Hirst M., Gilks B., Marra M.A., Huntsman D.G. ARID1A gene mutations in endometriosis associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S., Wang T.L., Shih I.M., Mao T.L., Nakayama K., Roden R., Glas R., Slamon D., Diaz L., Vogelstein B., Kinzler K.W., Velculescu V.E., Papadopoulos N. Exomic sequences of ovarian clear cell carcinomas. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reisman D., Glaros S., Thompson E.A. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 8.Medina P.P., Sanchez-Cespedes M. Involvement of the chromatin-remodeling factor BRG1/SMARCA4 in human cancer. Epigenetics. 2008;3:64–68. doi: 10.4161/epi.3.2.6153. [DOI] [PubMed] [Google Scholar]
- 9.Huang J., Zhao Y.L., Li Y., Fletcher J.A., Xiao S. Genomic and functional evidence for an ARID1A tumor suppressor role. Genes Chromosomes Cancer. 2007;46:745–750. doi: 10.1002/gcc.20459. [DOI] [PubMed] [Google Scholar]
- 10.Shih Ie M., Sheu J.J., Santillan A., Nakayama K., Yen M.J., Bristow R.E., Vang R., Parmigiani G., Kurman R.J., Trope C.G., Davidson B., Wang T.L. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci USA. 2005;102:14004–14009. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheu J.J., Choi J.H., Yildiz I., Tsai F.J., Shaul Y., Wang T.L., Shih Ie M. The roles of human sucrose nonfermenting protein 2 homologue in the tumor-promoting functions of Rsf-1. Cancer Res. 2008;68:4050–4057. doi: 10.1158/0008-5472.CAN-07-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loyola A., Huang J.Y., LeRoy G., Hu S., Wang Y.H., Donnelly R.J., Lane W.S., Lee S.C., Reinberg D. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol. 2003;23:6759–6768. doi: 10.1128/MCB.23.19.6759-6768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama K., Nakayama N., Jinawath N., Salani R., Kurman R.J., Shih Ie M., Wang T.L. Amplicon profiles in ovarian serous carcinomas. Int J Cancer. 2007;120:2613–2617. doi: 10.1002/ijc.22609. [DOI] [PubMed] [Google Scholar]
- 14.Schwab M. Amplification of oncogenes in human cancer cells. Bioessays. 1998;20:473–479. doi: 10.1002/(SICI)1521-1878(199806)20:6<473::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 15.Choi J.H., Sheu J.J., Guan B., Jinawath N., Markowski P., Wang T.L., Shih Ie M. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009;69:1407–1415. doi: 10.1158/0008-5472.CAN-08-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson B., Trope C.G., Wang T.L., Shih Ie M. Expression of the chromatin remodeling factor Rsf-1 is upregulated in ovarian carcinoma effusions and predicts poor survival. Gynecol Oncol. 2006;103:814–819. doi: 10.1016/j.ygyno.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 17.Mao T.L., Hsu C.Y., Yen M.J., Gilks B., Sheu J.J., Gabrielson E., Vang R., Cope L., Kurman R.J., Wang T.L., Shih Ie M. Expression of Rsf-1, a chromatin-remodeling gene, in ovarian and breast carcinoma. Hum Pathol. 2006;37:1169–1175. doi: 10.1016/j.humpath.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Maeda D., Chen X., Guan B., Nakagawa S., Yano T., Taketani Y., Fukayama M., Wang T.L., Shih Ie M. Rsf-1 (HBXAP) expression is associated with advanced stage and lymph node metastasis in ovarian clear cell carcinoma. Int J Gynecol Pathol. 2010;30:30–35. doi: 10.1097/PGP.0b013e3181e9a319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheu J.J., Hua C.H., Wan L., Lin Y.J., Lai M.T., Tseng H.C., Jinawath N., Tsai M.H., Chang N.W., Lin C.F., Lin C.C., Hsieh L.J., Wang T.L., Shih Ie M., Tsai F.J. Functional genomic analysis identified epidermal growth factor receptor activation as the most common genetic event in oral squamous cell carcinoma. Cancer Res. 2009;69:2568–2576. doi: 10.1158/0008-5472.CAN-08-3199. [DOI] [PubMed] [Google Scholar]
- 20.Petersen P.E. The World Oral Health Report 2003: continuous improvement of oral health in the 21st century–the approach of the WHO Global Oral Health Programme. Community Dent Oral Epidemiol. 2003;31(Suppl 1):3–23. doi: 10.1046/j..2003.com122.x. [DOI] [PubMed] [Google Scholar]
- 21.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–316. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Rousseau A., Lim M.S., Lin Z., Jordan R.C. Frequent cyclin D1 gene amplification and protein overexpression in oral epithelial dysplasias. Oral Oncol. 2001;37:268–275. doi: 10.1016/s1368-8375(00)00114-7. [DOI] [PubMed] [Google Scholar]
- 23.Freier K., Schwaenen C., Sticht C., Flechtenmacher C., Muhling J., Hofele C., Radlwimmer B., Lichter P., Joos S. Recurrent FGFR1 amplification and high FGFR1 protein expression in oral squamous cell carcinoma (OSCC) Oral Oncol. 2007;43:60–66. doi: 10.1016/j.oraloncology.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Rubin Grandis J., Melhem M.F., Gooding W.E., Day R., Holst V.A., Wagener M.M., Drenning S.D., Tweardy D.J. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90:824–832. doi: 10.1093/jnci/90.11.824. [DOI] [PubMed] [Google Scholar]
- 25.Toh Y., Nicolson G.L. The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications. Clin Exp Metastasis. 2009;26:215–227. doi: 10.1007/s10585-008-9233-8. [DOI] [PubMed] [Google Scholar]
- 26.Bochar D.A., Wang L., Beniya H., Kinev A., Xue Y., Lane W.S., Wang W., Kashanchi F., Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 27.Cheng S.W., Davies K.P., Yung E., Beltran R.J., Yu J., Kalpana G.V. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 28.Huang J.Y., Shen B.J., Tsai W.H., Lee S.C. Functional interaction between nuclear matrix-associated HBXAP and NF-kappaB. Exp Cell Res. 2004;298:133–143. doi: 10.1016/j.yexcr.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S., Young R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 30.Vignali M., Hassan A.H., Neely K.E., Workman J.L. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flanagan J.F., Peterson C.L. A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res. 1999;27:2022–2028. doi: 10.1093/nar/27.9.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varga-Weisz P. Chromatin remodeling factors and DNA replication. Prog Mol Subcell Biol. 2005;38:1–30. doi: 10.1007/3-540-27310-7_1. [DOI] [PubMed] [Google Scholar]
- 33.Wu S., Shi Y., Mulligan P., Gay F., Landry J., Liu H., Lu J., Qi H.H., Wang W., Nickoloff J.A., Wu C. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao A., Li H., Shechter D., Ahn S.H., Fabrizio L.A., Erdjument-Bromage H., Ishibe-Murakami S., Wang B., Tempst P., Hofmann K., Patel D.J., Elledge S.J., Allis C.D. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cosma M.P., Tanaka T., Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 36.Morettini S., Podhraski V., Lusser A. ATP-dependent chromatin remodeling enzymes and their various roles in cell cycle control. Front Biosci. 2008;13:5522–5532. doi: 10.2741/3096. [DOI] [PubMed] [Google Scholar]
- 37.Muchardt C., Yaniv M. When the SWI/SNF complex remodels.the cell cycle. Oncogene. 2001;20:3067–3075. doi: 10.1038/sj.onc.1204331. [DOI] [PubMed] [Google Scholar]
- 38.Ring H.Z., Vameghi-Meyers V., Wang W., Crabtree G.R., Francke U. Five SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin (SMARC) genes are dispersed in the human genome. Genomics. 1998;51:140–143. doi: 10.1006/geno.1998.5343. [DOI] [PubMed] [Google Scholar]
- 39.Klochendler-Yeivin A., Muchardt C., Yaniv M. SWI/SNF chromatin remodeling and cancer. Curr Opin Genet Dev. 2002;12:73–79. doi: 10.1016/s0959-437x(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 40.Lee D.W., Zhang K., Ning Z.Q., Raabe E.H., Tintner S., Wieland R., Wilkins B.J., Kim J.M., Blough R.I., Arceci R.J. Proliferation-associated SNF2-like gene (PASG): a SNF2 family member altered in leukemia. Cancer Res. 2000;60:3612–3622. [PubMed] [Google Scholar]
- 41.Sheu J.J., Guan B., Choi J.H., Lin A., Lee C.H., Hsiao Y.T., Wang T.L., Tsai F.J., Shih IeM. Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J Biol Chem. 2010;285:38260–38269. doi: 10.1074/jbc.M110.138735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoreactivity of anti-Rsf-1 in oral squamous cell carcinoma tissues and cells. A: Immunoreactivity was weak or undetectable in normal control and non-neoplastic tumor-adjacent epithelial cells. B: A stepwise and significant increment of Rsf-1 expression in non-neoplastic epithelium, CIS, and tumor cells could also be discerned.


