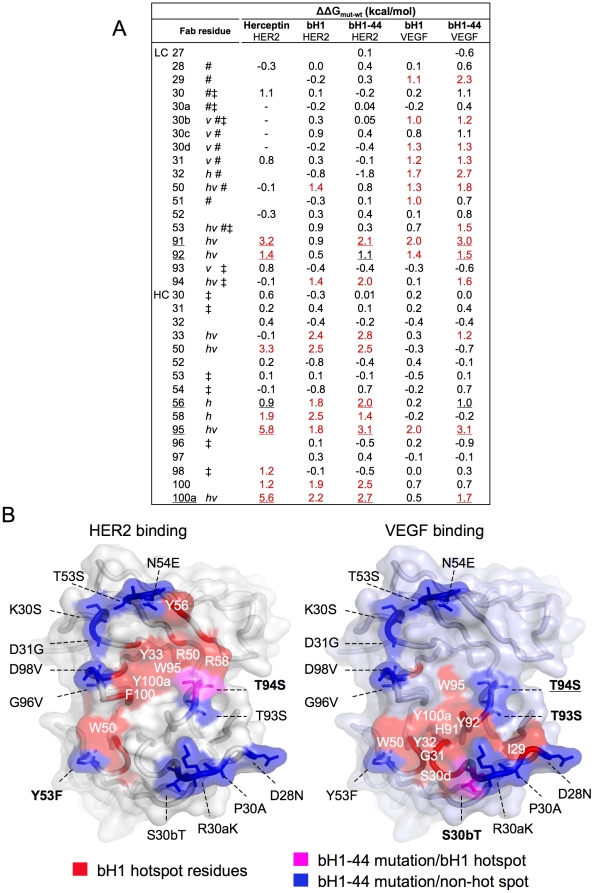
Figure 3. Mapping of the evolved dual specific interactions.
The changes in binding free energy (ΔΔGmut-wt) when a Fab residue is mutated to alanine (based on data from Kelley et. al. Biochemistry, 1993, Bostrom et. al., Science, 2009, and the current study) are shown (A). Amino acids are numbered as in Table 1 and denoted with h or v if making structural contact with HER2 or VEGF (within 4.5 Å), respectively, # if mutated from Herceptin to bH1, and ‡ if mutated from bH1 to bH1-44. Underlined residues are energetically important (ΔΔGmut-wt greater than ∼1 kcal/mol, also underlined) for Herceptin/HER2, bH1-44/HER2 and bH1-44/VEGF interactions indicating importance for all three interactions. Hotspot residues highlighted in red are those with ΔΔGmut-wt greater than ∼10% of the total binding free energy of each interaction. “-” denotes no residue in Herceptin at the position. Values not determined are left blank. (B) bH1-44 mutations (from bH1) are mapped to locate predominantly outside of functional hotspots and structural contact for HER2 binding (left) or VEGF binding (right) on the surface of the bH1 paratope modeled with side chains of bH-44. The residues are colored red if they are bH1 hotspot residues for HER2 or VEGF binding, blue if the residues are mutated from bH1 to bH1-44 and not part of bH1 hotspot for each respective interaction, or magenta if the mutated residues are part of bH1 hotspot. A mutation is bolded if the residue is a structural contact site in the respective bH1 interface or underlined if the mutated residue gains significant functional importance.