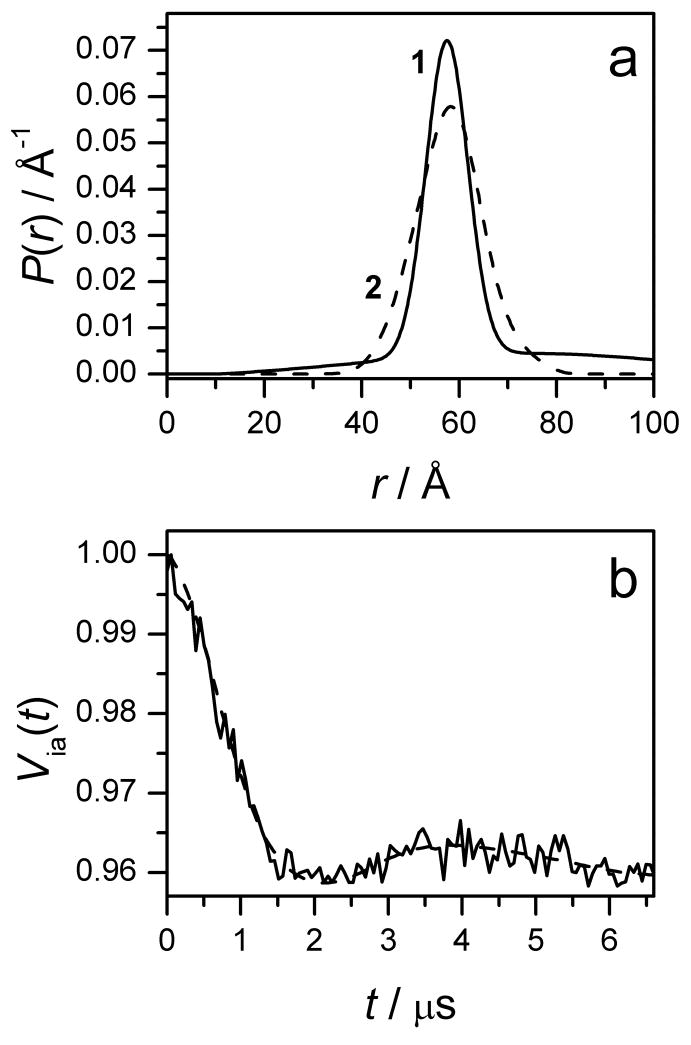
Figure 5.
(a) Trace 1, distance distribution function P(r) obtained from Via(t) of the double-stranded Gd595-DNA (average of kinetics 1 and 2 presented in Fig. 4b) using “the two-Gaussian fit option” of DeerAnalysys software package [78]. P(r) is the sum of two Gaussians, with (xo, δ) = (57.3 Å, 6 Å) and (73 Å, 50 Å), where xo and δ are the center and width of the Gaussians [78]. The ratio of weights (area integrals) for these Gaussians is 0.64:0.36. Trace 2, distance distribution function obtained by MD simulations. For this function x̄ = 57.6 Å and .
(b) Experimental (solid) and calculated (dashed) intrapair kinetics for the double-stranded Gd595-DNA. The calculated kinetics corresponds to the distribution function shown by trace 1 in Fig. 5a.