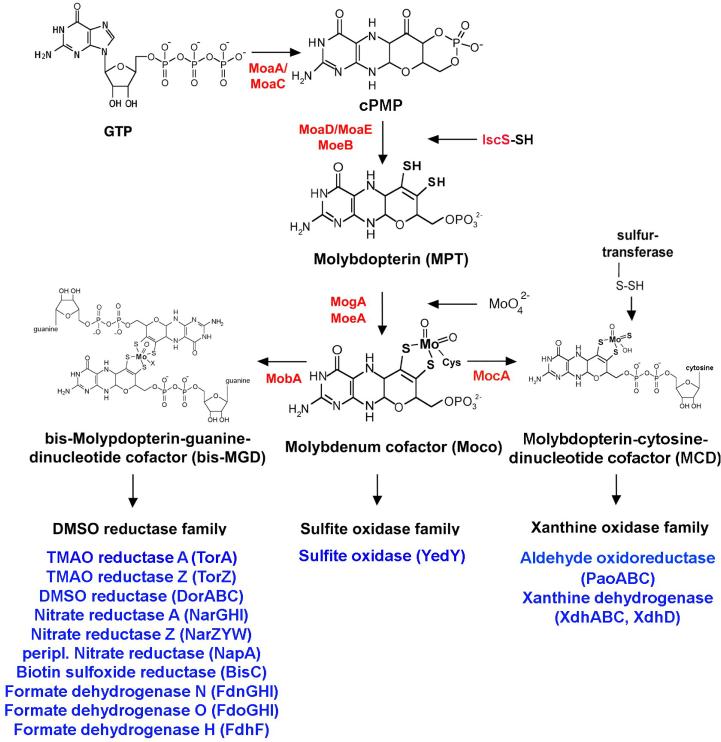
Figure 2. The biosynthesis of Moco in E. coli.
Shown is a scheme of the biosynthetic pathway for Moco biosynthesis in E. coli and the proteins involved in this pathway. Moco is formed from 5'GTP with Precursor Z and MPT as intermediates. The molybdoenzymes are classified into three different enzyme families, the DMSO reductase family, the sulfite oxidase family and the xanthine oxidase family, a categorization based on the coordination of the molybdenum center. For enzymes of the DMSO reductase family, Moco is further modified by the attachment of GMP to form MGD, and two equivalents of MGD are bound to a single molybdenum to form the bis-MGD variant of the cofactor. Additional molybdenum ligands can be an oxo- or a sulfido group in addition to a serine, a cysteine, a selenocysteine or a hydroxide and/or water molecule. For enzymes of the sulfite oxidase family, Moco is directly inserted without further addition of a nucleotide and coordinated by an additional cysteine ligand in the enzyme. For enzymes in the E. coli xanthine oxidase family, Moco is further modified by the addition of a cytosine nucleotide to form the MPT-cytosine dinucleotide (MCD) form of the cofactor. Additionally, a terminal sulfur ligand is added to the molybdenum site, generating sulfurated molybdenum MCD. An additional ligand at the Mo-center usually is a hydroxo-group. The names of the proteins involved in the reactions are colored in red, and the identified and characterized molybdoenzymes of each E. coli molybdoenzyme family are shown in blue.