Abstract
The provenance of white sharks (Carcharodon carcharias) in the Mediterranean is both a conundrum and an important conservation issue. Considering this species's propensity for natal philopatry, any evidence that the Mediterranean stock has little or no contemporary immigration from the Atlantic would suggest that it is extraordinarily vulnerable. To address this issue we sequenced the mitochondrial control region of four rare Mediterranean white sharks. Unexpectedly, the juvenile sequences were identical although collected at different locations and times, showing little genetic differentiation from Indo-Pacific lineages, but strong separation from geographically closer Atlantic/western Indian Ocean haplotypes. Historical long-distance dispersal (probably a consequence of navigational error during past climatic oscillations) and potential founder effects are invoked to explain the anomalous relationships of this isolated ‘sink’ population, highlighting the present vulnerability of its nursery grounds.
Keywords: Mediterranean, white shark, mitochondrial DNA, conservation, climate change, migration
1. Introduction
The movement of marine megafauna around the globe has significant and lasting consequences for ecosystems, especially where those species are top predators [1,2]. Some are known to undertake long transoceanic migrations [3–6], with an attendant risk of navigational error. Observations that water temperature appears to influence the movements of many marine species, including sharks [7,8], has led to the suggestion that thermal fronts may act as navigational cues during migration [9]. Such cues are easily disrupted during periods of climate change, producing anomalous distributions of some species [10]. Most of these instances probably go unnoticed, or remain little more than a historical anecdote, having no significant or lasting effect on the species' global distribution or on the ecosystem in which they become resident [2]. However, for species exhibiting natal female philopatry, such navigational errors may result in a founding population becoming closely associated with a new location, often outside the normal home range of the source population or well beyond the species' usual distribution. Once established as a top predator in a new location such founders may become effectively isolated, making them a vulnerable yet highly significant component of the ecosystem.
A wide-ranging species inhabiting sub-polar to tropical seas of both hemispheres, the great white shark, Carcharodon carcharias (Linneaus, 1758) has been documented in the Mediterranean [11,12]. The oldest white shark material preserved in Europe dates from 1640 to 1660; however, the capture date and locality are unknown [13]. Records of Mediterranean white sharks date back to the 1820–1850s, mainly from Italy [14] or Sicily [11]. However, the first legitimate scientific record of Mediterranean white sharks probably dates to 1901, when a 4.5 m female caught off the coast of Capo San Croce, Augusta, eastern Sicily was dissected, revealing three human corpses [15]. White sharks are recorded from all coasts of the Mediterranean western basin, most frequently the eastern side, with the most consistent reports in the Sicilian Channel and nearby waters. In the eastern basin, most observations are from the north, particularly the Adriatic, while the warmer, more saline south-eastern region returns infrequent records. Instances of large mature individuals, neonates and pregnant females in these waters [11,16,17] imply the existence of pupping and nursery grounds. However, there are no accounts of the origin and genetic profile of Mediterranean white sharks owing to rarity of samples.
The global phylogeography of C. carcharias is yet to be studied comprehensively. Nuclear gene flow throughout the Indian Ocean, and highly distinct mitochondrial haplotypes from populations either side of both the Indian and Pacific Oceans, suggest female philopatry and long-term isolation [18]. However, the evolutionary history of remaining disjunct populations is poorly known. Concordant with the hypothesis of natal philopatry, tagging studies reveal that although this species makes rapid and long transoceanic movements in both the Indian Ocean (a round trip of 11 100 km [3]) and northeast Pacific (traversing 2000–5000 km [19]), individuals adhere to a highly predictable cycle, persistently returning to natal coastal locations following migration [3,19,20], a behaviour which places populations at risk from local threats.
Evidence of dramatic population declines in large predatory sharks in the Mediterranean [21] and the potential consequences of trophic cascade [1] make it imperative to determine the utilization and connectivity of natural populations in vulnerable areas such as nursery/pupping grounds, especially in species exhibiting natal philopatry. Here, we report the first genetic analysis of Mediterranean white sharks, using the mtDNA control region. To elucidate the origins and relationships of rare Mediterranean white sharks to other stocks their haplotypes were compared with those from the north-eastern Pacific (NEP; California) [19], south-western Pacific (Australia, AU; and New Zealand, NZ), western Indian Ocean (South Africa, SA) [18] and the north-west Atlantic (NWA).
2. Material and methods
(a). Specimens
Mediterranean samples consisted of ethanol-preserved heart tissue from two neonate sharks caught off the coast of Altinoluk in the Bay of Edremit in the north-eastern Aegean Sea (eastern Mediterranean basin, Turkey) on 1 and 4 July 2008 (from separate parents [17]), a fin clip from a juvenile caught in Aras Dizra, Tunisia (also eastern Mediterranean basin) on 20 April 2006 [22], together with dried tissue from a shark caught in a tuna net off Favignana, Sicily (western Mediterranean basin) in the late 1980s. Additionally, two NWA fin clip samples (immature males, caught 1994) were collected off the west Florida coast, USA.
(b). Molecular methods and analysis
Genomic DNA was extracted by phenol–chloroform procedures, and the mtDNA control region amplified using primers and conditions from Pardini et al. [18]. Amplified products were purified using QIAquick (QIAGEN) columns and commercially sequenced. Work on the historical sample was undertaken in a shark-DNA-free laboratory, under sterile conditions, with tools and surfaces cleaned frequently. Blank samples were used to verify lack of contamination.
The six new sequences were aligned using CLUSTALX [23] against 49 previously published sequences [18,19] including animals sampled in the south-eastern Pacific (AU and NZ; AY026196–AY026209, AY026211), NEP (GU002321–GU002302) and western Indian Ocean (SA; AY026210, AY026212–AY026224). Additionally, two sequences from the porbeagle (Lamna nasus) were used as an outgroup. Measures of haplotype (h) and nucleotide (π) diversity were calculated with Arlequin v. 3.1.1 [24]. A median-joining network was constructed to depict haplotype relationships using NETWORK [25]. Based on likelihood-ratio tests (MODELTEST 3.7 [26]), the most appropriate model of evolution was the HKY + G model, with a gamma distribution parameter of 0.5332. Phylogenetic relationships were examined using MrBayes v. 3.1.2 [27]. Markov chain Monte Carlo (MCMC) simulations were run for 2 000 000 generations, and the first 10 per cent less-optimal tree generations were discarded as ‘burn-in’. PhyML v. 3.0 [28] was also used to calculate the maximum-likelihood tree. An alternative tree topology (NWA/SA as sister clade to the Mediterranean) was compared with the ML inferred tree using the Shimodaira–Hasegawa (SH) test, as executed in PAUP* v. 4.0b10 [29] with RELL approximations and 1000 bootstrap replicates to produce a null distribution of differences in log-likelihoods. The SH test compares the difference in log-likelihoods of competing tree topologies. Finally, evolutionary rates for the control region were calculated in two steps: initially, the nucleotide substitutions per site (Da) were calculated between sequences from the Pacific (AU, NZ, NEP) versus either the Atlantic (NWA) or eastern Indian ocean (SA) using DNASP v. 5.10 [30], and calibrated with either the rise of the Panama Isthmus (3.5 Ma [31]) or Sunda-Sahul shelves (5 Ma [32]; the most protracted and, for an epipelagic species, the most significant lineage-splitting period of sea level drop early in white shark evolution).
3. Results
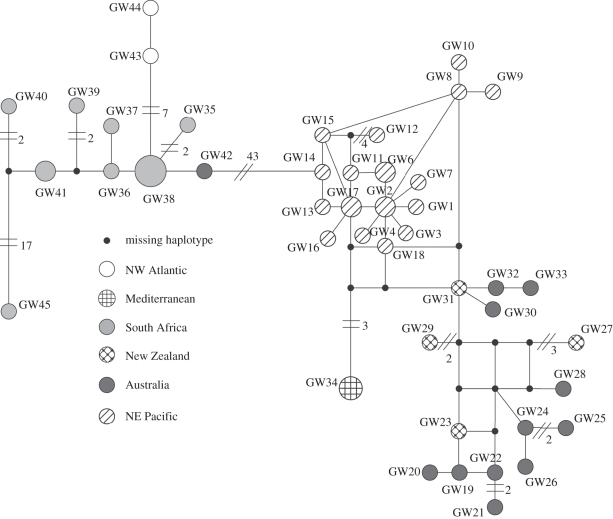
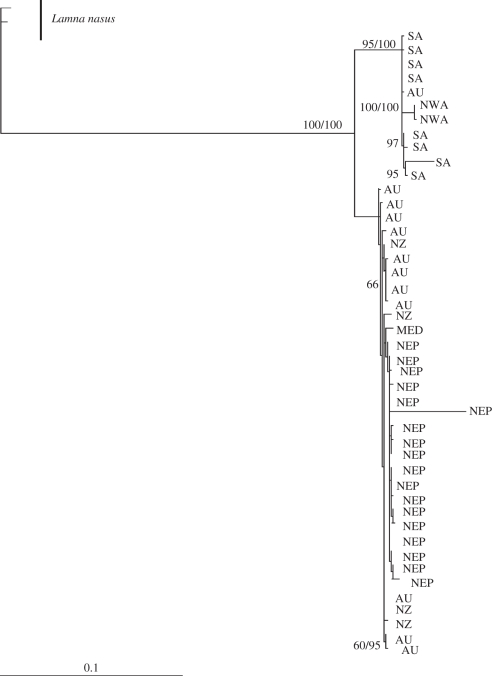
A 1083 bp sequence of the mtDNA control region was obtained for five of the new samples (three Mediterranean and two NWA; accession nos HQ540294–HQ540298). Overall, 95 polymorphic sites revealed 45 haplotypes (figure 1) from 54 sequences examined, showing that C. carcharias exhibits the highest haplotype (0.9888 ± 0.0075) and nucleotide (0.0223 ± 0.0110) diversities of any shark studied to date [33–36]. Summary statistics for each population are given in electronic supplementary material, table S1. Yet remarkably, three of the four Mediterranean (MED) samples shared the same haplotype (GW34), whereas each NWA sample was unique (GW43, GW44). The median-joining network indicates that the MED haplotypes show little genetic differentiation from Indo-Pacific sharks, with only five mutational steps separating them from either NEP (GW18 and GW17) or NZ (GW31; figure 1), and six steps from AU (GW30 and GW32) haplotypes. Partial sequences (297 bp; accession nos HQ540299–HQ540300) from the Sicilian skin sample also gave a unique haplotype, but was separated by only three hypothetical mutational steps from an NEP haplotype (electronic supplementary material, figure S1). Mediterranean white sharks from European museum collections [14] are presently inaccessible for destructive analysis. However, it is unlikely that full sequences would be obtained from this material. The South African haplotypes form a distinct group separated from the NWA by only seven mutational steps. However, the SA/NWA group differs from the Indo-Pacific/MED group by 43 mutations. Both Bayesian and ML analyses corroborate the close genetic similarity of the Mediterranean and western Indo-Pacific Ocean haplotypes (figure 2). Further, SH tests of an alternative topology to the ML inferred tree (difference in ln L = 32.36, p < 0.003) firmly rejected the NWA/SA as sister clade to the Mediterranean. Evolutionary rates for the white shark control region were calculated as either 1.19 or 0.74 per cent divergence between lineages per million years, calibrated with the rise of the Panama Isthmus or the Sunda-Sahul shelves, respectively. This corresponds to a Mediterranean–Indo-Pacific population divergence of 348–565 Ka. Significantly, this dates divergence of the Mediterranean stock to the Late Pleistocene, a period of extreme dynamic climatic and eustatic events, suggested to have dramatic impacts on the distribution and size of fish populations.
Figure 1.
A median-joining network of 45 mtDNA control region haplotypes from white shark sampling areas, illustrating the close affinity of Mediterranean samples with the Indo-Pacific clade. Circle size is proportional to the frequency of the haplotype. Unless specified, single mutational steps are assumed between haplotypes; line breaks with a number indicate the mutational steps.
Figure 2.
Maximum-likelihood phylogeny, depicting genetic relationships among C. carcharias, inferred from mtDNA control region sequences (an alignment length of 1087 bp including indels). The two main haplotype lineages correspond to the Indo-Pacific and South African/north-west Atlantic clades. Mediterranean samples cluster with Indo-Pacific haplotypes, from which they are only weakly differentiated. The tree is rooted with Lamna nasus and values above branches indicate support for each node based on Bayesian/ML inference. Bootstrap values under 60% are not shown.
4. Discussion
Analysis of the mitochondrial control region of several rare Mediterranean white sharks suggests that they have little genetic variability and a close affinity with the Indo-Pacific clade. Current understanding of white shark biology suggests recent Lessepsian migrations [37], characteristic of other Indo-Pacific taxa inhabiting the eastern Mediterranean, are an unlikely explanation. Here, we argue that this highly vulnerable stock may owe its origins to a historical anomalous migratory event, rather than persistent historical or contemporary dispersal via the Atlantic.
The close affinity of the MED and Indo-Pacific (figure 2) dismisses founding of the Mediterranean population solely by a simple dispersal scenario from the geographically closest populations of the NWA (separated by more than 51 mutational steps; figure 1). Consequently, the question arises as to whether the five or six mutations differentiating MED and Indo-Pacific haplotypes indicate an isolated Mediterranean subpopulation, or a stock with contemporary and persistent demographic connections with the Indo-Pacific. The most divergent haplotypes within either the south-western Pacific or the NEP differ by nine (GW21 to either GW27 or GW33) and eight substitutions (GW12 to the group of GW18/7/4/3/1), respectively. This suggests that divergence between Indo-Pacific and MED haplotypes corresponds to levels of genetic differentiation observed within each group. However, the large geographical distance separating Mediterranean and Oceania waters, and the absence of the Mediterranean haplotype along routes from the Indo-Pacific, including comprehensively sampled South African locations, suggest that demographically contiguous contemporary or historical populations are unlikely.
The shortest route connecting Indo-Pacific and MED populations is through the Red Sea and Suez Canal. Nevertheless, occurrence of white sharks in the Red Sea is disputed [38], and a recent record from the Arabian Sea is considered a misidentification [39]. Zuffa et al. [40] reported the presence of C. carcharias off southern and western Madagascar, with northernmost records off Kenya and Zanzibar, findings consistent with descriptions of a 6.4 m pregnant female off Kenya [41]. Additionally, the thermal barrier of tropical waters, and hypersalinity of the Red Sea, Bitter Lakes and Suez Canal, were thought to prevent white shark dispersal. However, satellite tagging reveals that this species tolerates a wide temperature range [3], consistent with it traversing the Red Sea. Nevertheless, Fergusson [11] suggests some seasonality of white sharks, catch data indicating movement into cooler northern areas (the Adriatic) from the south (Tunisia and Sicilian Channel) in summer months. Consistent with this, high summer sea surface temperature (SST) of approximately 26°C (close to the species's upper limit [42]) in the Red Sea and adjacent waters, and a lack of confirmed sightings, suggests this area acts as a thermal barrier, making a Lessepsian migration route less likely. So, what mechanism can be invoked to explain the anomalous presence of these haplotypes? Consideration of when the Mediterranean and Indo-Pacific sequences diverged may help elucidate this issue.
The lower estimate for white shark control region mutation rate is similar to that of scalloped hammerheads (0.8% [43]) and lemon sharks (0.67% [35]), corroborating the slow evolution of elasmobranchs [44]. Hence, the MED/Indo-Pacific separation is estimated to have occurred in the Late Pleistocene (approx. 0.45 Ma), roughly corresponding with the postulated origin of the NEP population [19] (with the caveat that calculations are based upon a single, unconserved region). Significantly, the dynamic eustatic and climatic changes of this period have been implicated in promoting dramatic changes in population dynamics and range fluctuations [45]. A recognized yet infrequent historical connection between the Indo-Pacific, southwest Atlantic [46] and ultimately the Mediterranean open intermittently at this time has been used to explain the distribution of other pelagic fish stocks [47]. This alternative longer route evokes eustatic regression events, widely accepted to produce vicariance in pelagic species, entailing historical dispersal across the Indian Ocean to South Africa, into the Atlantic and north to the Straits of Gibraltar. Glacial events over the last 700 000 years have caused repeated sea level falls of more than 80 m below present heights. Notably, on such occasions the Sunda and Sahul shelves formed a barrier between the Pacific and Indian Oceans, with vigorous leakage of Indian Ocean fauna later facilitated by an enhanced Agulhas current during Pleistocene inter-glacials [46]. This oceanic interchange has been suggested to account for the distribution of Pacific clades of swordfish in the eastern Atlantic and particularly the Mediterranean, which they entered during an inter-glacial, surviving the last glacial maximum (LGM) in an eastern refuge [47]. It could be argued that swordfish and white sharks exhibit ecological commonalities in their colonization of the Mediterranean. Both have similar temperature tolerances and natal philopatry [3,18], and swordfish are also prey of white sharks. However, though Alvarado-Bremer et al. [47] suggest that some Mediterranean swordfish haplotypes are of Pacific origin, they emphasize that the ubiquity and contemporary presence of these haplotypes in the southern Atlantic is consistent with sustained unidirectional gene flow. Furthermore, despite evidence that the Mediterranean stock survived the LGM isolated in the eastern Mediterranean, their relationship with Pacific haplotypes in the Atlantic suggests that glacial conditions did not erase the earlier signature of persistent migration into the Atlantic and subsequently the Mediterranean.
Although several alternative hypotheses can be invoked to explain white shark colonization of the Mediterranean, every scenario except historical infrequent long-distance dispersal relies upon evidence of ubiquitous clades in the Atlantic and Pacific, whereas the global population is composed of clades specific to ocean basins. There is no recorded co-occurrence of clades in the Atlantic, as is often cited for other pelagic species whose analogous phylogeographic patterns are explained by protracted unidirectional gene flow from the Indo-Pacific into the Atlantic [47]. Nor is there evidence of Pacific lineages in the southern Atlantic, which would also support such a scenario. Additionally, palaeoclimatic reconstructions of Pleistocene Atlantic SST [48] do not support retreat from the Atlantic to leave a relictual Mediterranean population during glacial maxima; temperatures remained above the critical 15°C threshold for white shark movements in the Atlantic [49].
It would therefore seem a less than parsimonious explanation to consider the GWS Pacific stock once ubiquitous but eradicated from the Atlantic during glacial maxima. Conversely, glacial conditions promoting southward expansion of Arctic fauna may have sustained rather than displaced white sharks in the equatorial Atlantic. Rather, assuming that in situ divergence corresponds to an inter-glacial around Marine Isotope Stages 12 to 10, we suggest the estimated divergence time of Mediterranean and Pacific haplotypes at around 450 Kya. Hence, historical accident during dramatic and dynamic Pleistocene climate change can be invoked to explain Mediterranean white shark origins, at least partially. This scenario would be consistent with dominance of few unique Pacific lineage haplotypes across the eastern Mediterranean. A later migratory event following MED/Indo-Pacific divergence cannot be ruled out, but could be invoked by the same climatic trigger occurring repeatedly in the late Pleistocene.
The climatic instability of the Pleistocene may have induced navigational errors, with sharks following an Agulhas ring or eddy. During inter-glacials of the last 700 000 years this would have been remarkably stronger than the contemporary phenomenon [46], directing animals north along the African coast. Following expanding swordfish and bluefin tuna populations, which arrived via these anomalous currents, a propensity to swim east to natal grounds would have led ultimately to entrapment in the Mediterranean. Entrapment is also proposed to account for the strong Indo-Pacific monophyletic mitochondrial signature of Mediterranean swordfish [47]. Dominance of few distinct haplotypes over such a wide area would be wholly consistent with isolation, population reduction, natal philopatry and restriction to eastern Mediterranean refugia during glacial advances. Even if white sharks were able to penetrate the cooler Atlantic at this time, natal philopatry, as evidenced by our juvenile samples, would invariably associate these haplotypes with the Mediterranean. Genetic differences are also apparent between northern Atlantic and Mediterranean populations of other transoceanic dispersing marine predators, such as the sperm whale, Physeter macrocephalus [5]. Their differentiation can be explained by female philopatry, but their colonization is clearly attributable to a founder event involving northern Atlantic individuals. Like Lessepsian migrants, founding white sharks may have encountered oceanographic characteristics and prey availability in the eastern (as distinct from the western) Mediterranean basin approximating their Indo-Pacific natal areas. This scenario, consistent with the absence of these Mediterranean haplotypes from our comprehensive genetic analyses of South African white sharks [18,50], argues for a disjunct distribution of the Indo-Pacific clade.
As a species exhibiting strong natal female philopatry, these sharks probably constitute a sink population established at a time of extreme climate change. Consequently, if the haplotypes recovered truly reflect white shark diversity there would seem little potential for replacement females to enter and establish in the Mediterranean should the current population become unsustainable. Some migration of white sharks into the Mediterranean cannot be dismissed, but they are rare in the north-eastern Atlantic [51], with most captures from oceanic islands, particularly the Azores, of mature individuals. An occasional visitor [52], its origins and limits of distribution in this region remain poorly known [11]. Additionally, no information is available from the Straits of Gibraltar, where interchange between the Atlantic and the Mediterranean would be most apparent. In contrast, available sightings suggest that large individuals are resident at eastern basin Mediterranean sites for a year or more [53], with catches year round. Hence, migration between these populations, while not impossible, seems unlikely. Currently, there is no available tagging data for Mediterranean white sharks, and recent analysis of a few tagged north-western Atlantic individuals indicates migration along the US coast (Cape Cod, 2009; G. Skomal 2010, personal communication) rather than across the Atlantic. Indeed, white sharks are encountered most frequently in continental shelf waters from Cape Cod to Cape Hatteras in the north-western Atlantic Ocean [49], showing a more seasonal (January–April) presence west of Florida [54]. Nevertheless, even if males from the Atlantic occasionally mate with Mediterranean females, the natal philopatry of the latter guarantees that continuity of the Mediterranean stock remains inextricably linked to the fate of these anomalous haplotypes. Future analyses incorporating both nuclear and mitochondrial markers in larger population samples will determine the likelihood of male- and female-mediated gene flow into the Mediterranean, determining if this is truly a closed population established by infrequent historical migration of ‘founder’ females.
A key member of a vulnerable ecosystem, currently of huge commercial and scientific value, the Mediterranean white shark population may be considered a consequence of historical accident and female philopatry, revealing it to be at greater risk of local extirpations than previously thought. Demise of this top predator in a relatively small and contained sea may precipitate a catastrophic trophic cascade, already recorded in more robust ecosystems following depletion of populations of large sharks [1]. It is disturbing to speculate about the disproportionate deleterious effects white shark loss may have on ecosystem stability in this relatively enclosed and economically important environment.
Acknowledgements
This work was supported by Aberdeen University, the Marine Biological Association, Boğaziçi University and the Scientific and Technological Research Council of Turkey (TÜBİTAK). We are also grateful to Eleonora de Sabata (MedSharks, Italy) for supplying a sample, Nicolas Pade for porbeagle sequences, Colin Macleod and two anonymous referees for helpful comments. In memoriam: Dave Noble (d. 20 October 2010)—always and forever a support and inspiration.
References
- 1.Myers R. A., Baum J. K., Shepherd T. D., Powers S. P., Peterson C. H. 2007. Cascading effects of the loss of apex predatory sharks from a coastal ocean. Science 315, 1846–1850 10.1126/science.1138657 (doi:10.1126/science.1138657) [DOI] [PubMed] [Google Scholar]
- 2.Crowder L., Norse E. 2008. Essential ecological insights for marine ecosystem-based management and marine spatial planning. Mar. Policy 32, 772–778 10.1016/j.marpol.2008.03.012 (doi:10.1016/j.marpol.2008.03.012) [DOI] [Google Scholar]
- 3.Bónfil R., Meÿer M., Scholl M. C., Johnson R., O'Brien S., Oosthuizen H., Swanson S., Kotze D., Paterson M. 2005. Transoceanic migration, spatial dynamics, and population linkages of white sharks. Science 310, 100–103 10.1126/science.1114898 (doi:10.1126/science.1114898) [DOI] [PubMed] [Google Scholar]
- 4.Gore M. A., Rowat D., Hall J., Gell F. R., Ormond R. F. 2008. Transatlantic migration and deep mid-ocean diving by basking shark. Biol. Lett. 4, 395–398 10.1098/rsbl.2008.0147 (doi:10.1098/rsbl.2008.0147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelhaupt D., et al. 2009. Female philopatry in coastal basins and male dispersion across the North Atlantic in a highly mobile marine species, the sperm whale (Physeter macrocephalus). Mol. Ecol. 18, 4193–4205 10.1111/j.1365-294X.2009.04355.x (doi:10.1111/j.1365-294X.2009.04355.x) [DOI] [PubMed] [Google Scholar]
- 6.Skomal G. B., Zeeman S. I., Chisholm J. H., Summers E. L., Walsh H. J., McMahon K. W., Thorrold S. R. 2009. Transequatorial migrations by basking sharks in the Western Atlantic Ocean. Curr. Biol. 19, 1019–1022 10.1016/j.cub.2009.04.019 (doi:10.1016/j.cub.2009.04.019) [DOI] [PubMed] [Google Scholar]
- 7.Holts D. B., Bedford D. W. 1993. Horizontal and vertical movements of the shortfin mako shark, Isurus oxyrinchus, in the southern California Bight. Aust. J. Mar. Freshw. Res. 44, 901–909 10.1071/MF9930901 (doi:10.1071/MF9930901) [DOI] [Google Scholar]
- 8.Wilson S. G., Polovina J. J., Stewart B. S., Meekan M. G. 2006. Movements of whale sharks (Rhincodon typus) tagged at Ningaloo Reef, Western Australia. Mar. Biol. 148, 1157–1166 10.1007/s00227-005-0153-8 (doi:10.1007/s00227-005-0153-8) [DOI] [Google Scholar]
- 9.Sherrill-Mix S. A., James M. C., Myers R. A. 2008. Migration cues and timing in leatherback sea turtles. Behav. Ecol. 19, 231–236 10.1093/beheco/arm104 (doi:10.1093/beheco/arm104) [DOI] [Google Scholar]
- 10.Corten A. 2001. Northern distribution of North Sea herring as a response to high water temperature and/or low food abundance. Fish. Res. 50, 189–204 10.1016/S0165-7836(00)00251-4 (doi:10.1016/S0165-7836(00)00251-4) [DOI] [Google Scholar]
- 11.Fergusson I. K. 1996. Distribution and autoecology of the white shark in the Eastern North Atlantic and the Mediterranean Sea. In Great white sharks: the biology of Carcharodon carcharias (eds Klimley A. P., Ainley D. G.), pp. 321–345 San Diego, CA: Academic Press [Google Scholar]
- 12.Kabasakal H., Kabasakal E. 2004. Sharks captured by commercial fishing vessels off the coast of Turkey in the northern Aegean Sea. Ann. Ser. Hist. Nat. 14, 171–180 [Google Scholar]
- 13.De Maddalena A. 2005. The great white shark, Carcharodon carcharias (Linnaeus, 1758) of the Settala Museum, Milan. Bollettino del Museo Civico di Storia Naturale di Venezia 57, 149–154 [Google Scholar]
- 14.De Maddalena A. 2006. A catalogue of great white sharks Carcharodon carcharias (Linnaeus, 1758) preserved in European museums. J. Natl Mus., Nat. History Ser. 175, 109–125 [Google Scholar]
- 15.Condorelli M., Perrando G. C. 1909. Notizie sul Carcharcdon carcharias L., catturato nelle acque di Augusta e considerazioni medico-legali su resti umani trovati nel suo tubo digerente. Soc. Zool. Ital. Boll. 1909, 164–183 [Google Scholar]
- 16.Saidi B., Bradai M. N., Bouaïn A., Guélorget O., Capepé C. 2005. Capture of a pregnant female white shark, Carcharodon carcharias (Lamnidae) in the Gulf of Gabès (southern Tunisia, central Mediterranean) with comments on oophagy in sharks. Cybium 29, 303–307 [Google Scholar]
- 17.Kabasakal H., Gedikoğlu S. 2008. Two new-born great white sharks, Carcharodon carcharias (Linnaeus, 1758) (Lamniformes; Lamnidae) from Turkish waters of the north Aegean Sea. Acta Adriat. 49, 125–135 [Google Scholar]
- 18.Pardini A. T., et al. 2001. Sex-biased dispersal of great white sharks. Nature 412, 139–140 10.1038/35084125 (doi:10.1038/35084125) [DOI] [PubMed] [Google Scholar]
- 19.Jorgensen S., et al. 2009. Philopatry and migration of Pacific white sharks. Proc. R. Soc. B 277, 679–688 10.1098/rspb.2009.1155 (doi:10.1098/rspb.2009.1155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasby-Lucas N., Dewar H., Lam C. H., Goldman K. J., Domeier M. L., Bograd S. J. 2009. White shark offshore habitat: a behavioral and environmental characterization of the eastern Pacific shared offshore foraging area. PLoS ONE 4, e8163. 10.1371/journal.pone.0008163 (doi:10.1371/journal.pone.0008163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferretti F., Myers R. A., Serena F., Lotze H. K. 2008. Loss of large predatory sharks from the Mediterranean Sea. Conserv. Biol. 22, 952–964 10.1111/j.1523-1739.2008.00938.x (doi:10.1111/j.1523-1739.2008.00938.x) [DOI] [PubMed] [Google Scholar]
- 22.Iglésias S. P. 2009. Chondrichthyans from the North-eastern Atlantic and the Mediterranean (a natural classification based on collection specimens). Provisional version 03, internet prepublication, 1 November 2009. See http://www.mnhn.fr/iccanam
- 23.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Excoffier L., Laval G., Schneider S. 2005. Arlequin v. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- 25.Bandelt H. J., Forster P., Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 [DOI] [PubMed] [Google Scholar]
- 26.Posada D., Crandall K. A. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818 10.1093/bioinformatics/14.9.817 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 27.Huelsenbeck J. P., Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 [DOI] [PubMed] [Google Scholar]
- 28.Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 [DOI] [PubMed] [Google Scholar]
- 29.Swofford D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4.0b10 Sunderland, MA: Sinauer Associates [Google Scholar]
- 30.Librado P., Rozas J. 2009. DNASP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 10.1093/bioinformatics/btp187 (doi:10.1093/bioinformatics/btp187) [DOI] [PubMed] [Google Scholar]
- 31.Coates A. G., Jackson J. B. C., Collins L. S., Cronin T. M., Dowsett H. J., Bybell L. M., Jung P., Obando J. A. 1992. Closure of the Isthmus of Panama; the near-shore marine record of Costa Rica and western Panama. Geol. Soc. Am. Bull. 104, 814–828 (doi:10.1130/0016-7606(1992)104<0814:COTIOP>2.3.CO;2) [DOI] [Google Scholar]
- 32.Haq B. U., Hardenbol J., Vail P. R. 1987. Chronology of fluctuating sea levels since the Triassic. Science 235, 1156–1167 10.1126/science.235.4793.1156 (doi:10.1126/science.235.4793.1156) [DOI] [PubMed] [Google Scholar]
- 33.Hoelzel A. R., Shivji M. S., Magnussen J., Francis M. P. 2006. Low worldwide genetic diversity in the basking shark (Cetorhinus maximus). Biol. Lett. 2, 639–642 10.1098/rsbl.2006.0513 (doi:10.1098/rsbl.2006.0513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castro A. L. F., Stewart B. S., Wilson S. G., Hueter R. E., Meekan M. G., Motta P. J., Bowen B. W., Karl S. A. 2007. Population genetic structure of earth's largest fish, the whale shark (Rhincodon typus). Mol. Ecol. 16, 5183–5192 10.1111/j.1365-294X.2007.03597.x (doi:10.1111/j.1365-294X.2007.03597.x) [DOI] [PubMed] [Google Scholar]
- 35.Schultz J. K., Feldheim K. A., Gruber S. H., Ashley M. V., McGovern T. M., Bowen B. W. 2008. Global phylogeography and seascape genetics of the lemon sharks (genus Negaprion). Mol. Ecol. 17, 5336–5348 10.1111/j.1365-294X.2008.04000.x (doi:10.1111/j.1365-294X.2008.04000.x) [DOI] [PubMed] [Google Scholar]
- 36.Portnoy D. S., McDowell J. R., Heist E. J., Musick J. A., Graves J. E. 2010. World phylogeography and male-mediated gene flow in the sandbar shark, Carcharhinus plumbeus. Mol. Ecol. 19, 1994–2010 10.1111/j.1365-294X.2010.04626.x (doi:10.1111/j.1365-294X.2010.04626.x) [DOI] [PubMed] [Google Scholar]
- 37.Por F. D. 1971. One hundred years of Suez Canal—a century of Lessepsian migration: retrospect and viewpoints. Syst. Zool. 20, 138–159 10.2307/2412054 (doi:10.2307/2412054) [DOI] [Google Scholar]
- 38.Bónfil R., Abdallah M. 2004. Field identification guide to the sharks and rays of the Red Sea and Gulf of Aden. In FAO species identification guide for fishery purposes. Rome, Italy: FAO [Google Scholar]
- 39.Moore A. B. M., Compagno L. J. V., Fergusson I. K. 2007. The Persian/Arabian Gulf's sole great white shark Carcharodon carcharias (Lamniformes: Lamnidae) record from Kuwait: misidentification of a sandtiger shark Carcharias taurus (Lamniformes: Odontaspididae). Zootaxa 1591, 67–68 [Google Scholar]
- 40.Zuffa M., Van Grevelynghe G., De Maddalena A., Storai T. 2002. Records of the white shark, Carcharodon carcharias (Linnaeus, 1758), from the western Indian Ocean. S. Afr. J. Sci. 98, 347–349 [Google Scholar]
- 41.Cliff G., Compagno L. J. V., Smale M. J., van der Elst R. P., Wintner S. P. 2000. First records of white sharks, Carcharodon carcharias, from Mauritius, Zanzibar, Madagascar, and Kenya. S. Afr. J. Sci. 96, 365–367 [Google Scholar]
- 42.Cliff G., Dudley S. F. J., Davis B. 1989. Sharks caught in the protective gill nets off Natal, South Africa. 2. The great white shark Carcharodon carcharias (Linnaeus). S. Afr. J. Mar. Sci. 8, 131–144 [Google Scholar]
- 43.Duncan K. M., Martin A. P., Bowen B. W., De Couet H. G. 2006. Global phylogeography of the scalloped hammerhead shark (Sphyrna lewini). Mol. Ecol. 15, 2239–2251 10.1111/j.1365-294X.2006.02933.x (doi:10.1111/j.1365-294X.2006.02933.x) [DOI] [PubMed] [Google Scholar]
- 44.Martin A. P., Naylor G. J. P., Palumbi S. R. 1992. Rates of mitochondrial DNA evolution in sharks are slow compared with mammals. Nature 357, 153–155 10.1038/357153a0 (doi:10.1038/357153a0) [DOI] [PubMed] [Google Scholar]
- 45.Hofreiter M., Stewart J. 2009. Ecological change, range fluctuations and population dynamics during the Pleistocene. Curr. Biol. 19, R584–R594 10.1016/j.cub.2009.06.030 (doi:10.1016/j.cub.2009.06.030) [DOI] [PubMed] [Google Scholar]
- 46.Peeters F. J. C., Acheson R., Brummer G.-J. A., de Ruijter W. P. M., Schneider R. R., Ganssen G. M., Ufkes E., Kroon D. 2004. Vigorous exchange between the Indian and Atlantic oceans at the end of the past five glacial periods. Nature 430, 661–665 10.1038/nature02785 (doi:10.1038/nature02785) [DOI] [PubMed] [Google Scholar]
- 47.Alvarado Bremer J., Vinas J., Mejuto J., Ely B., Pla C. 2005. Comparative phylogeography of Atlantic bluefin tuna and swordfish: the combined effects of vicariance, secondary contact, introgression, and population expansion on the regional phylogenies of two highly migratory pelagic fishes. Mol. Phylogenet. Evol. 36, 169–187 10.1016/j.ympev.2004.12.011 (doi:10.1016/j.ympev.2004.12.011) [DOI] [PubMed] [Google Scholar]
- 48.Schefuss E., Sinninghe Damste J. S., Jansen J. H. F. 2004. Forcing of tropical Atlantic sea surface temperatures during the mid-Pleistocene transition. Paleoceanography 19, A4029 P 10.1029/2003PA000892 (doi:10.1029/2003PA000892) [DOI] [Google Scholar]
- 49.Casey J. C., Pratt H. L., Jr 1985. Distribution of the white shark, Carcharodon carcharias, in the western North Atlantic. South. Calif. Acad. Sci. Mem. 9, 2–14 [Google Scholar]
- 50.Gubili C. 2008. Application of molecular genetics for conservation of the great white shark Carcharodon carcharias, L. 1758. PhD thesis, University of Aberdeen, UK [Google Scholar]
- 51.Quero C. 1984. Lamnidae. In Fishes of the north-eastern Atlantic and the Mediterranean, vol. 1 (eds Whitehead P. J. P., Bauchot M. L., Hureau J. C., Nielsen J., Tortonese E.), pp. 33–38 Paris, France: UN Environmental Science Organization [Google Scholar]
- 52.Springer S. 1979. Carcharodon carcharias (Linnaeus). In Checklist of the fishes of the north-eastern Atlantic and of the Mediteranean, vol. 1 (eds Hureau J. C., Monod T.). Paris, France: CLOFNAM, UNESCO Press [Google Scholar]
- 53.Bini G. 1960. Attacco documentato di pescecane Carcharodon carcharias. Bol. Pesca. Pisciolt. Idribiol. Ann. 15, 136–139 [Google Scholar]
- 54.Adams D. H., Mitchell M. E., Parsons G. R. 1994. Seasonal occurrence of the White Shark, Carcharodon carcharias, in waters off the Florida west coast, with notes on its life history. Mar. Fish. Rev. 56, 24–28 [Google Scholar]