Figure 4.
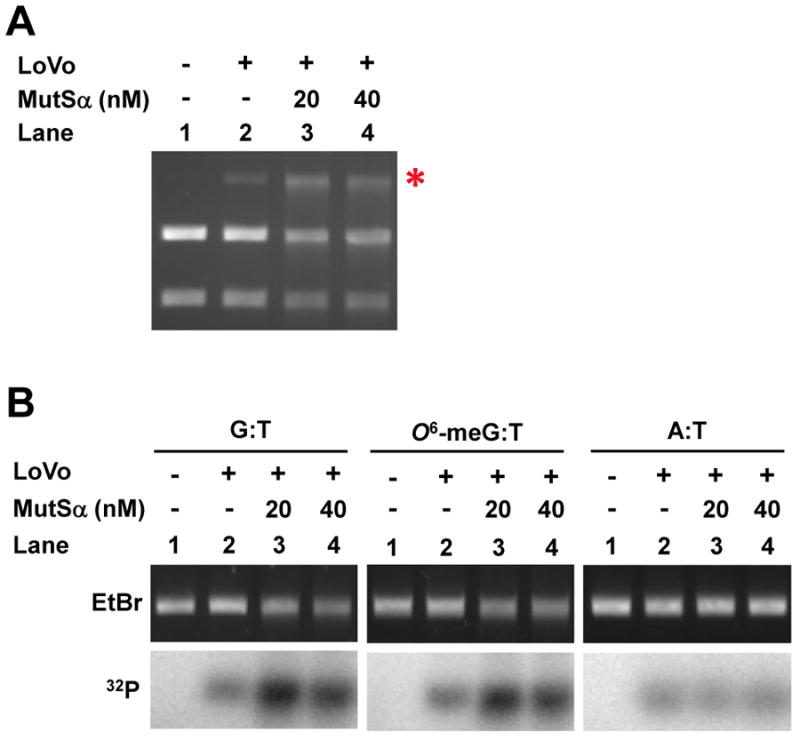
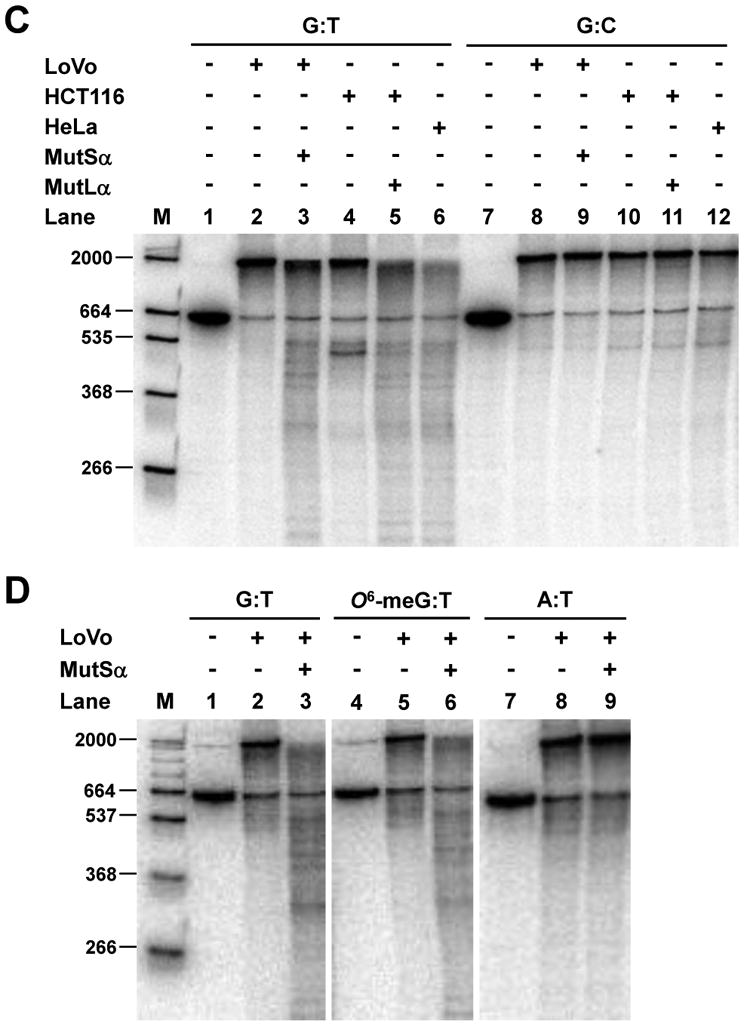
Nick-directed mismatch-provoked excision. (A) Excision assay measured by restriction endonuclease sensitivity was performed with MSH2-deficient LoVo nuclear extract and pSCW01_GT substrate in the absence of exogenous dNTPs. Recovered DNA was digested with BamHI & AseI endonucleases and separated by electrophoresis. Red asterisk denotes BamHI-resistant gapped DNA after excision. (B) Excision assay measured by annealing of an oligonucleotide probe using pSCW02_GT, pSCW02_O6meGT, and pSCW02 homoduplex substrates. Gapped DNA was digested with AseI, and annealed to a 32P-oligonucleotide probe that spans the mismatch site. (C) Excision assay utilizing Southern blotting was with pSCW01_GT DNA substrate. Nicked homoduplex pSCW01 was used to measure ExoI random excision. Lane 1 and 7 are controls with nicked pSCW01_GT or pSCW01 in the absence of nuclear extract. (D) Experiments were performed as in (C) but with the pSCW02_GT, pSCW02_O6-meGT and pSCW02 homoduplex substrates in a Southern blotting assay.
