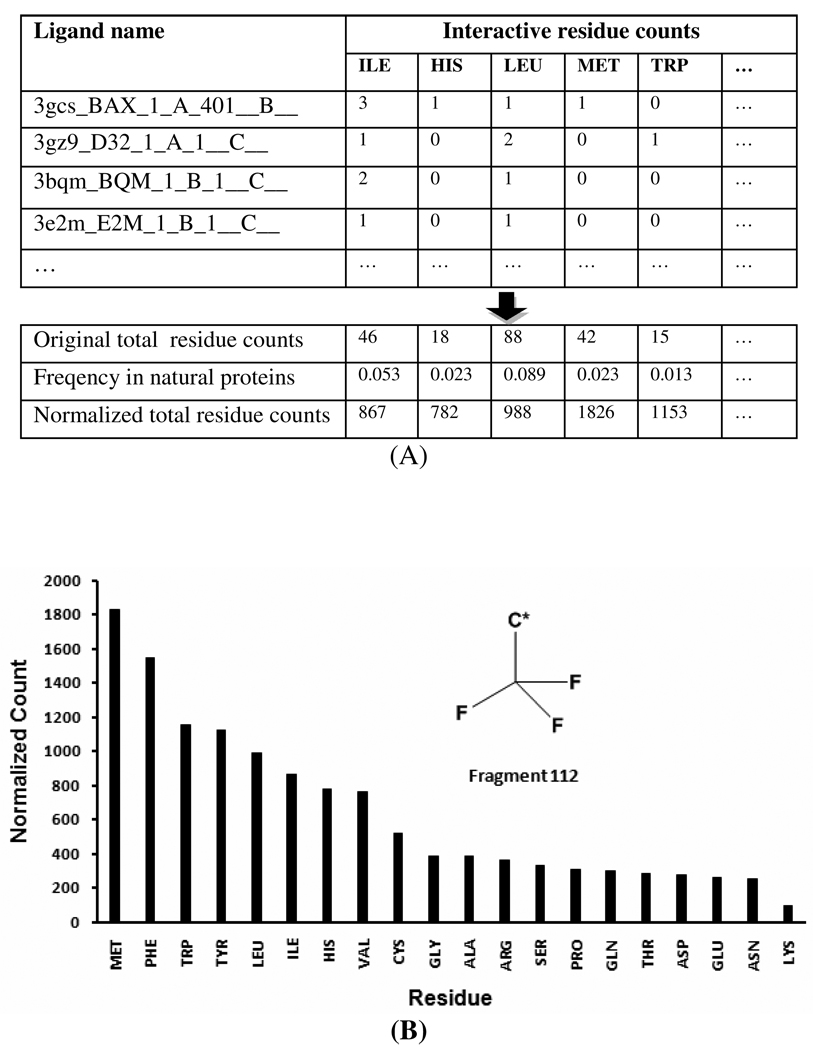
Figure 3. An interaction profile is mapped out between the fragment 112 and 20 amino acid residues.
A: The interactive residues for fragment 112 (trifluoromethyl) in the non-redundant PDB structures are counted according to residues names, and the residue counts are then normalized by the correspondent residue frequencies in natural proteins. B. The graph plot of the interaction profile (normalized residues counts) for fragment 112. This interaction profile shows that the top five residues (MET, PHE, TRP, TYR and LEU) are the most favored five residues whereas THR, ASP, GLU, ASN and LYS are the least favored five residues for fragment 112.