Abstract
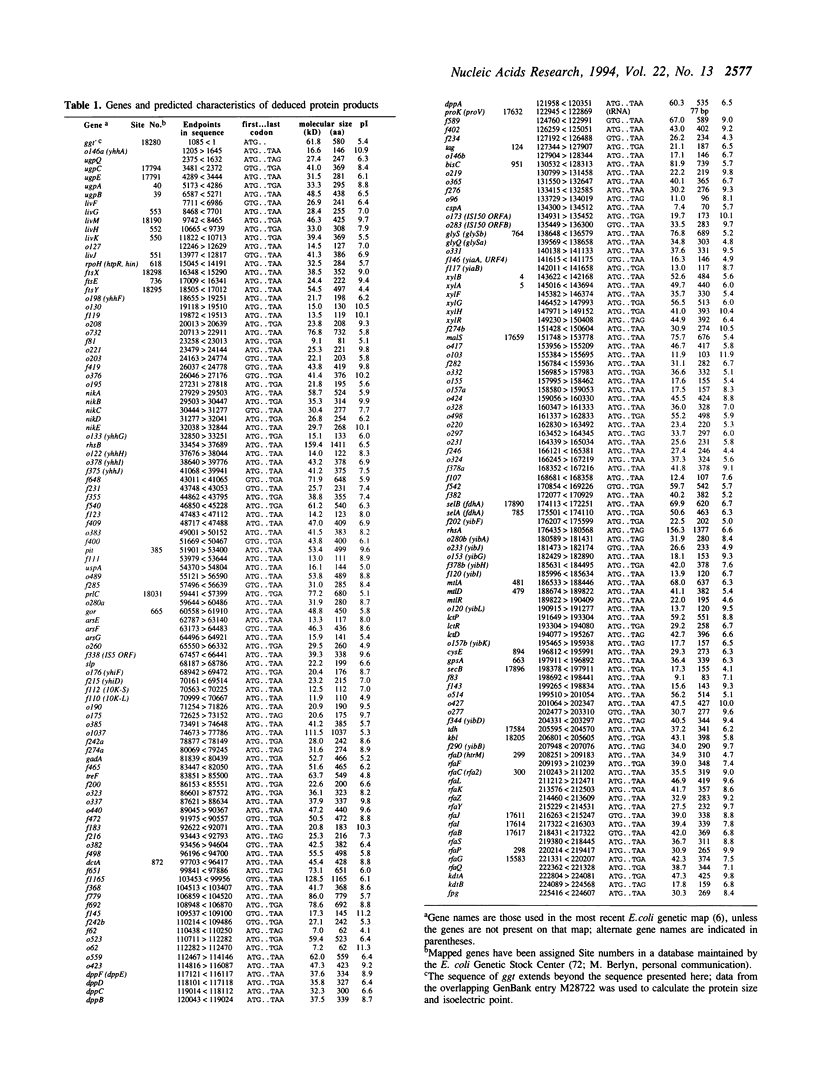
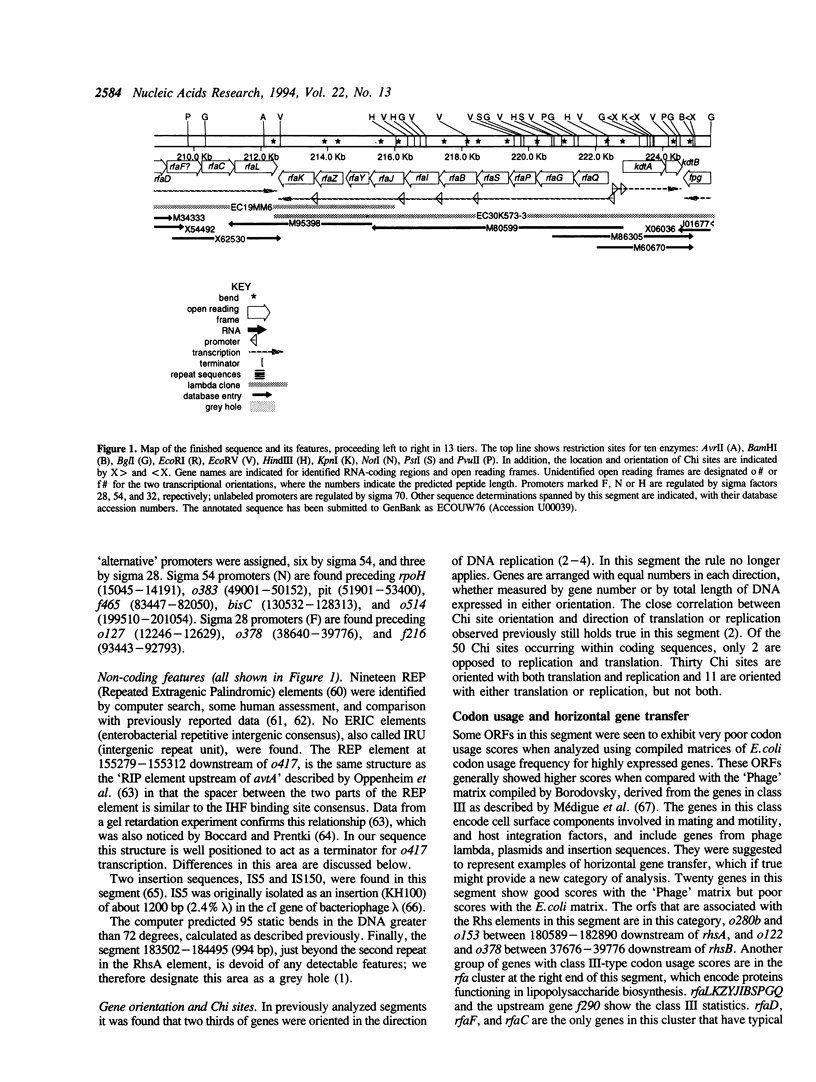
The DNA sequence of a 225.4 kilobase segment of the Escherichia coli K-12 genome is described here, from 76.0 to 81.5 minutes on the genetic map. This brings the total of contiguous sequence from the E.coli genome project to 725.1 kb (76.0 to 92.8 minutes). We found 191 putative coding genes (ORFs) of which 72 genes were previously known, and 110 of which remain unidentified despite literature and similarity searches. Seven new genes--arsE, arsF, arsG, treF, xylR, xylG, and xylH--were identified as well as the previously mapped pit and dctA genes. The arrangement of proposed genes relative to possible promoters and terminators suggests 90 potential transcription units. Other features include 19 REP elements, 95 computer-predicted bends, 50 Chi sites, and one grey hole. Thirty-one putative signal peptides were found, including those of thirteen known membrane or periplasmic proteins. One tRNA gene (proK) and two insertion sequences (IS5 and IS150) are located in this segment. The genes in this region are organized with equal numbers oriented with or against replication.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abouhamad W. N., Manson M., Gibson M. M., Higgins C. F. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol Microbiol. 1991 May;5(5):1035–1047. doi: 10.1111/j.1365-2958.1991.tb01876.x. [DOI] [PubMed] [Google Scholar]
- Adams M. D., Wagner L. M., Graddis T. J., Landick R., Antonucci T. K., Gibson A. L., Oxender D. L. Nucleotide sequence and genetic characterization reveal six essential genes for the LIV-I and LS transport systems of Escherichia coli. J Biol Chem. 1990 Jul 15;265(20):11436–11443. [PubMed] [Google Scholar]
- Atkinson E. M., Long S. R. Homology of Rhizobium meliloti NodC to polysaccharide polymerizing enzymes. Mol Plant Microbe Interact. 1992 Sep-Oct;5(5):439–442. doi: 10.1094/mpmi-5-439. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn M. B., Letovsky S. Genome-related datasets within the E. coli Genetic Stock Center database. Nucleic Acids Res. 1992 Dec 11;20(23):6143–6151. doi: 10.1093/nar/20.23.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl R. P., Vielmetter W. Complete maps of IS1, IS2, IS3, IS4, IS5, IS30 and IS150 locations in Escherichia coli K12. Mol Gen Genet. 1989 Dec;220(1):147–153. doi: 10.1007/BF00260869. [DOI] [PubMed] [Google Scholar]
- Blattner F. R., Burland V., Plunkett G., 3rd, Sofia H. J., Daniels D. L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 1993 Nov 25;21(23):5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F. R., Fiandt M., Hass K. K., Twose P. A., Szybalski W. Deletions and insertions in the immunity region of coliphage lambda: revised measurement of the promoter-startpoint distance. Virology. 1974 Dec;62(2):458–471. doi: 10.1016/0042-6822(74)90407-3. [DOI] [PubMed] [Google Scholar]
- Boccard F., Prentki P. Specific interaction of IHF with RIBs, a class of bacterial repetitive DNA elements located at the 3' end of transcription units. EMBO J. 1993 Dec 15;12(13):5019–5027. doi: 10.1002/j.1460-2075.1993.tb06195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos W., Ehmann U., Forkl H., Klein W., Rimmele M., Postma P. Trehalose transport and metabolism in Escherichia coli. J Bacteriol. 1990 Jun;172(6):3450–3461. doi: 10.1128/jb.172.6.3450-3461.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993 Jun;16(3):551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- Conlin C. A., Miller C. G. Location of the prlC (opdA) gene on the physical map of Escherichia coli. J Bacteriol. 1993 Sep;175(17):5731–5732. doi: 10.1128/jb.175.17.5731-5732.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet J., Cauchois L., Blanche F., Debussche L., Thibaut D., Rouyez M. C., Rigault S., Mayaux J. F., Cameron B. Nucleotide sequence of a Pseudomonas denitrificans 5.4-kilobase DNA fragment containing five cob genes and identification of structural genes encoding S-adenosyl-L-methionine: uroporphyrinogen III methyltransferase and cobyrinic acid a,c-diamide synthase. J Bacteriol. 1990 Oct;172(10):5968–5979. doi: 10.1128/jb.172.10.5968-5979.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- David J. D., Wiesmeyer H. Control of xylose metabolism in Escherichia coli. Biochim Biophys Acta. 1970 Mar 24;201(3):497–499. doi: 10.1016/0304-4165(70)90171-6. [DOI] [PubMed] [Google Scholar]
- Dimri G. P., Rudd K. E., Morgan M. K., Bayat H., Ames G. F. Physical mapping of repetitive extragenic palindromic sequences in Escherichia coli and phylogenetic distribution among Escherichia coli strains and other enteric bacteria. J Bacteriol. 1992 Jul;174(14):4583–4593. doi: 10.1128/jb.174.14.4583-4593.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J. M., Taylor J. S., Latour D. J., Iuchi S., Lin E. C. Three overlapping lct genes involved in L-lactate utilization by Escherichia coli. J Bacteriol. 1993 Oct;175(20):6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundlieb S., Boos W. Alpha-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J Biol Chem. 1986 Feb 25;261(6):2946–2953. [PubMed] [Google Scholar]
- Fuchs R. MacPattern: protein pattern searching on the Apple Macintosh. Comput Appl Biosci. 1991 Jan;7(1):105–106. doi: 10.1093/bioinformatics/7.1.105. [DOI] [PubMed] [Google Scholar]
- Gendron N., Breton R., Champagne N., Lapointe J. Adenylosuccinate lyase of Bacillus subtilis regulates the activity of the glutamyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5389–5392. doi: 10.1073/pnas.89.12.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Saurin W., Perrin D., Bachellier S., Hofnung M. Palindromic units are part of a new bacterial interspersed mosaic element (BIME). Nucleic Acids Res. 1991 Apr 11;19(7):1375–1383. doi: 10.1093/nar/19.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J., Pollitt N. S., Inouye M. Major cold shock protein of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Jan;87(1):283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer S., Perham R. N. Glutathione reductase from Escherichia coli: cloning and sequence analysis of the gene and relationship to other flavoprotein disulfide oxidoreductases. Biochemistry. 1986 May 6;25(9):2736–2742. doi: 10.1021/bi00357a069. [DOI] [PubMed] [Google Scholar]
- Henrissat B., Claeyssens M., Tomme P., Lemesle L., Mornon J. P. Cellulase families revealed by hydrophobic cluster analysis. Gene. 1989 Sep 1;81(1):83–95. doi: 10.1016/0378-1119(89)90339-9. [DOI] [PubMed] [Google Scholar]
- Jahn D. Complex formation between glutamyl-tRNA synthetase and glutamyl-tRNA reductase during the tRNA-dependent synthesis of 5-aminolevulinic acid in Chlamydomonas reinhardtii. FEBS Lett. 1992 Dec 7;314(1):77–80. doi: 10.1016/0014-5793(92)81465-x. [DOI] [PubMed] [Google Scholar]
- Ji G., Silver S. Regulation and expression of the arsenic resistance operon from Staphylococcus aureus plasmid pI258. J Bacteriol. 1992 Jun;174(11):3684–3694. doi: 10.1128/jb.174.11.3684-3694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J. R., Henrich B., Plapp R. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol Gen Genet. 1991 Nov;230(1-2):230–240. doi: 10.1007/BF00290673. [DOI] [PubMed] [Google Scholar]
- Klein J. R., Plapp R. Locations of the envCD genes on the physical map of the Escherichia coli chromosome. J Bacteriol. 1992 Jun;174(11):3828–3829. doi: 10.1128/jb.174.11.3828-3829.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klena J. D., Pradel E., Schnaitman C. A. Comparison of lipopolysaccharide biosynthesis genes rfaK, rfaL, rfaY, and rfaZ of Escherichia coli K-12 and Salmonella typhimurium. J Bacteriol. 1992 Jul;174(14):4746–4752. doi: 10.1128/jb.174.14.4746-4752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J., Delcuve G. Thermosensitive mutants of Escherichia coli K-12 altered in the catalytic Subunit and in a Regulatory factor of the glutamy-transfer ribonucleic acid synthetase. J Bacteriol. 1975 May;122(2):352–358. doi: 10.1128/jb.122.2.352-358.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J., Söll D. Glutamyl transfer ribonucleic acid synthetase of Escherichia coli. 3. Influence of the 46K protein on the affinity of the 56K glutamyl transfer ribonucleic acid synthetase for its substrates. J Biol Chem. 1972 Aug 25;247(16):4982–4985. [PubMed] [Google Scholar]
- Liu L., Whalen W., Das A., Berg C. M. Rapid sequencing of cloned DNA using a transposon for bidirectional priming: sequence of the Escherichia coli K-12 avtA gene. Nucleic Acids Res. 1987 Nov 25;15(22):9461–9469. doi: 10.1093/nar/15.22.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T. C., Bewick M. A. The molecular mechanisms of dicarboxylic acid transport in Escherichia coli K12. The role and orientation of the two membrane-bound dicarboxylate binding proteins. J Biol Chem. 1978 Nov 10;253(21):7826–7831. [PubMed] [Google Scholar]
- Ma D., Cook D. N., Alberti M., Pon N. G., Nikaido H., Hearst J. E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993 Oct;175(19):6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallinder P. R., Pritchard A., Moir A. Cloning and characterization of a gene from Bacillus stearothermophilus var. non-diastaticus encoding a glycerol dehydrogenase. Gene. 1992 Jan 2;110(1):9–16. doi: 10.1016/0378-1119(92)90438-u. [DOI] [PubMed] [Google Scholar]
- Manson M. D., Blank V., Brade G., Higgins C. F. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature. 1986 May 15;321(6067):253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- Mayer R., Ross P., Weinhouse H., Amikam D., Volman G., Ohana P., Calhoon R. D., Wong H. C., Emerick A. W., Benziman M. Polypeptide composition of bacterial cyclic diguanylic acid-dependent cellulose synthase and the occurrence of immunologically crossreacting proteins in higher plants. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5472–5476. doi: 10.1073/pnas.88.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgola E. J., Adelberg E. A. Mutants of Escherichia coli K-12 with an altered glutamyl-transfer ribonucleic acid synthetase. J Bacteriol. 1970 Jul;103(1):178–183. doi: 10.1128/jb.103.1.178-183.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médigue C., Rouxel T., Vigier P., Hénaut A., Danchin A. Evidence for horizontal gene transfer in Escherichia coli speciation. J Mol Biol. 1991 Dec 20;222(4):851–856. doi: 10.1016/0022-2836(91)90575-q. [DOI] [PubMed] [Google Scholar]
- Navarro C., Wu L. F., Mandrand-Berthelot M. A. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol Microbiol. 1993 Sep;9(6):1181–1191. doi: 10.1111/j.1365-2958.1993.tb01247.x. [DOI] [PubMed] [Google Scholar]
- Oden K. L., Gladysheva T. B., Rosen B. P. Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol Microbiol. 1994 Apr;12(2):301–306. doi: 10.1111/j.1365-2958.1994.tb01018.x. [DOI] [PubMed] [Google Scholar]
- Oppenheim A. B., Rudd K. E., Mendelson I., Teff D. Integration host factor binds to a unique class of complex repetitive extragenic DNA sequences in Escherichia coli. Mol Microbiol. 1993 Oct;10(1):113–122. doi: 10.1111/j.1365-2958.1993.tb00908.x. [DOI] [PubMed] [Google Scholar]
- Osorio A. V., Camarena L., Salazar G., Noll-Louzada M., Bastarrachea F. Nitrogen regulation in an Escherichia coli strain with a temperature sensitive glutamyl-tRNA synthetase. Mol Gen Genet. 1993 Jun;239(3):400–408. doi: 10.1007/BF00276938. [DOI] [PubMed] [Google Scholar]
- Overduin P., Boos W., Tommassen J. Nucleotide sequence of the ugp genes of Escherichia coli K-12: homology to the maltose system. Mol Microbiol. 1988 Nov;2(6):767–775. doi: 10.1111/j.1365-2958.1988.tb00088.x. [DOI] [PubMed] [Google Scholar]
- Plunkett G., 3rd, Burland V., Daniels D. L., Blattner F. R. Analysis of the Escherichia coli genome. III. DNA sequence of the region from 87.2 to 89.2 minutes. Nucleic Acids Res. 1993 Jul 25;21(15):3391–3398. doi: 10.1093/nar/21.15.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyssegur J., Stoeber F. Genetic control of the 2-keto-3-deoxy-d-gluconate metabolism in Escherichia coli K-12: kdg regulon. J Bacteriol. 1974 Feb;117(2):641–651. doi: 10.1128/jb.117.2.641-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priefert H., Krüger N., Jendrossek D., Schmidt B., Steinbüchel A. Identification and molecular characterization of the gene coding for acetaldehyde dehydrogenase II (acoD) of Alcaligenes eutrophus. J Bacteriol. 1992 Feb;174(3):899–907. doi: 10.1128/jb.174.3.899-907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmanian M., Claus D. R., Oxender D. L. Multiplicity of leucine transport systems in Escherichia coli K-12. J Bacteriol. 1973 Dec;116(3):1258–1266. doi: 10.1128/jb.116.3.1258-1266.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina S., Georgopoulos C. The htrM gene, whose product is essential for Escherichia coli viability only at elevated temperatures, is identical to the rfaD gene. Nucleic Acids Res. 1991 Jul 25;19(14):3811–3819. doi: 10.1093/nar/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodolakis A., Casse F., Starka J. Morphological mutants of Escherichia coli K12. Mapping of the env C mutation. Mol Gen Genet. 1974 May 21;130(2):177–181. doi: 10.1007/BF00269088. [DOI] [PubMed] [Google Scholar]
- Rosenfeld S. A., Stevis P. E., Ho N. W. Cloning and characterization of the xyl genes from Escherichia coli. Mol Gen Genet. 1984;194(3):410–415. doi: 10.1007/BF00425552. [DOI] [PubMed] [Google Scholar]
- Saxena I. M., Lin F. C., Brown R. M., Jr Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol Biol. 1990 Nov;15(5):673–683. doi: 10.1007/BF00016118. [DOI] [PubMed] [Google Scholar]
- Saxena I. M., Lin F. C., Brown R. M., Jr Identification of a new gene in an operon for cellulose biosynthesis in Acetobacter xylinum. Plant Mol Biol. 1991 Jun;16(6):947–954. doi: 10.1007/BF00016067. [DOI] [PubMed] [Google Scholar]
- Schnaitman C. A., Parker C. T., Klena J. D., Pradel E. L., Pearson N. B., Sanderson K. E., MacClachlan P. R. Physical maps of the rfa loci of Escherichia coli K-12 and Salmonella typhimurium. J Bacteriol. 1991 Dec;173(23):7410–7411. doi: 10.1128/jb.173.23.7410-7411.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. G., Hamblin M. J., Kelly D. J. Purification, characterization and nucleotide sequence of the periplasmic C4-dicarboxylate-binding protein (DctP) from Rhodobacter capsulatus. Mol Microbiol. 1991 Dec;5(12):3055–3062. doi: 10.1111/j.1365-2958.1991.tb01865.x. [DOI] [PubMed] [Google Scholar]
- Silver S., Ji G., Bröer S., Dey S., Dou D., Rosen B. P. Orphan enzyme or patriarch of a new tribe: the arsenic resistance ATPase of bacterial plasmids. Mol Microbiol. 1993 May;8(4):637–642. doi: 10.1111/j.1365-2958.1993.tb01607.x. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Bell R. M., Cronan J. E., Jr A mutant of Escherichia coli auxotrophic for organic phosphates: evidence for two defects in inorganic phosphate transport. Mol Gen Genet. 1975 Dec 30;143(1):71–77. doi: 10.1007/BF00269422. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Kumagai H., Echigo T., Tochikura T. DNA sequence of the Escherichia coli K-12 gamma-glutamyltranspeptidase gene, ggt. J Bacteriol. 1989 Sep;171(9):5169–5172. doi: 10.1128/jb.171.9.5169-5172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommassen J., Eiglmeier K., Cole S. T., Overduin P., Larson T. J., Boos W. Characterization of two genes, glpQ and ugpQ, encoding glycerophosphoryl diester phosphodiesterases of Escherichia coli. Mol Gen Genet. 1991 Apr;226(1-2):321–327. doi: 10.1007/BF00273621. [DOI] [PubMed] [Google Scholar]
- Wang M. D., Liu L., Wang B. M., Berg C. M. Cloning and characterization of the Escherichia coli K-12 alanine-valine transaminase (avtA) gene. J Bacteriol. 1987 Sep;169(9):4228–4234. doi: 10.1128/jb.169.9.4228-4234.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C., Fear A. L., Calhoon R. D., Eichinger G. H., Mayer R., Amikam D., Benziman M., Gelfand D. H., Meade J. H., Emerick A. W. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8130–8134. doi: 10.1073/pnas.87.20.8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Ueguchi C., Yamada H., Mizuno T. Function of the Escherichia coli nucleoid protein, H-NS: molecular analysis of a subset of proteins whose expression is enhanced in a hns deletion mutant. Mol Gen Genet. 1993 Feb;237(1-2):113–122. doi: 10.1007/BF00282791. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]


