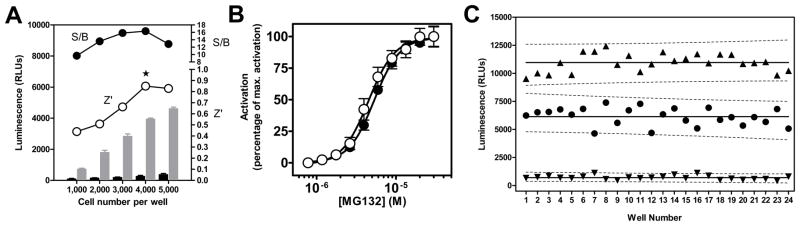
FIG. 2.
Wee1 degradation assay optimization and validation. (A) Cell seeding density optimization. After transfection with the K328M-Wee1-Luc contruct, HeLa cells were seeded at different densities ranging from 1,000 to 5,000 cells per well. Cells were either treated with DMSO alone (black bars) or with 30 μM of reference compounds MG132 (grey bars) for 20 hours. Z′ (○) and signal-to-background ratio (□) calculated based on relative luminescence unit (RLU) values between DMSO and MG132-treated cells are shown for each tested cell density. The star indicates the selected optimal cell density. Error bars represent the S.D. of each test condition (n=4). (B) Concentration-dependent activation of the K328M-Wee1-Luc reporter upon MG132 treatment in HeLa cells. Results in 1536-well plate format protocol using non-frozen cells (○) or frozen cells (●). Twenty-five nanoliters of a 10-point, 3:2 serial dilution starting at 30 μM were dispensed into an assay plate using a PinTool. Each data point is mean ± S.D. (n=16). The calculated EC50 was 4.54 ± 0.44 μM. (C) Scatter diagram of the controls of a representative 1536-well screening plate. Each plate contains three different controls: wells treated with DMSO only, noted as Low Controls (▼), wells receiving 30 μM of the reference compound MG132 (▲) and wells receiving a concentration of MG132 equivalent to its EC50, i.e. 5 μM (●). The calculated signal-to-background ratio was 15.53 and the Z′ factor 0.71. Solid lines represent the average luminescence value for each control type and the dashed lines the associated 95% prediction interval. Note the absence of overlap between the prediction intervals for all three control types.