Figure 1.
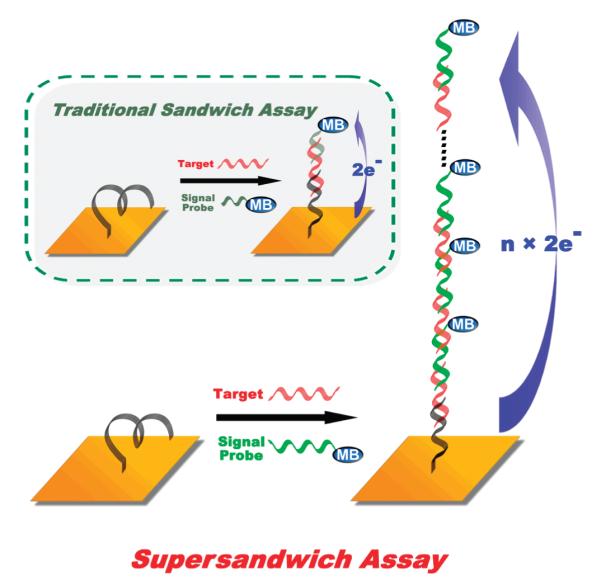
In a traditional DNA sandwich assay, a single target molecule hybridizes with a single signal probe (inset). Here we have modified this approach by using signal probes that hybridize to two regions of the target DNA, thus creating long concatamers containing multiple target molecules and signal probes and leading to improved signaling and detection limits. In the sensor design, the capture probe DNA and the signal probe (supersandwich) DNA are actually of the same sequence except for the modifications on the two ends. Of note, the affinity of the signal probe for the target is slightly higher than the affinity of the target for the capture probe (due to steric interactions with the electrode), and thus every time a capture probe is occupied, it is occupied by a sandwich and not simply by a target probe lacking a signaling probe.
