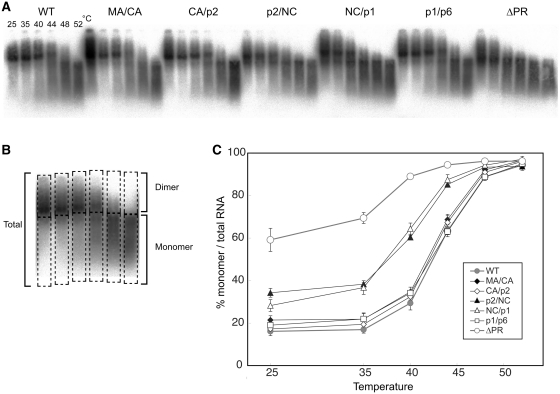
Figure 2.
Thermal stability of Viral RNA dimers of single Gag-cleavage site mutants. (A) Virion RNA profiles detected by northern blotting in a native agarose gel. Viruses were prepared by transfection of 293T cells with the wild-type (WT; pNLNh) virus or its derivative mutants. Aliquots of RNA extracted from virions were resuspended in T-buffer and incubated for 10 min in parallel reactions. The temperatures in which aliquots were incubated are indicated for each lane. (B) A schematic figure for dimer/monomer calculation. Each lane of the blot was separated into two parts, dimer or larger and monomer or smaller molecules as shown in boxes with dashed lines. The relative amounts of monomeric and total RNAs in each lane were quantitated with ImageGauge, and the percentage of the total represented by monomeric RNA was calculated for each RNA sample. (C) Thermal dissociation kinetics of RNA dimers. Results are the average of three independent experiments. Error bars represent SEM.