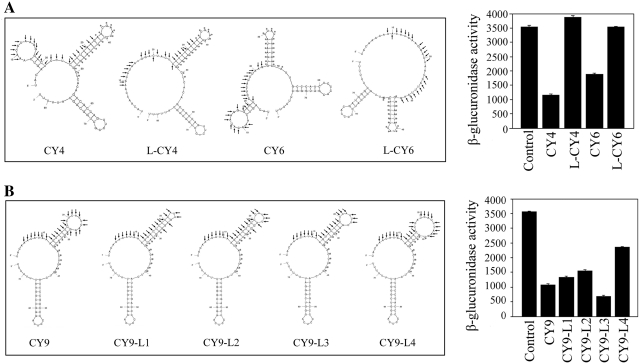
Figure 3.
Secondary structure and function of atsRNA. (A) The predicted secondary structures and effect of atsRNA mutants L-CY4 and L-CY6 designed based on CY4 and CY6, respectively. Compared with the corresponding atsRNAs, L-CY4 and L-CY6 lost the stem loop of mRNA base pairing region in secondary structure. (B) The predicted secondary structures and effect of atsRNA mutants designed based on CY9. CY9-L1, CY9-L2, CY9-L3 and CY9-L4 were CY9 mutants with different number of unpaired nucleotides in the loop structure of mRNA base pairing region. The sequences of predicted pairing region (indicated with arrows) and Hfq binding site (indicated with asterisks) of atsRNA are marked. The secondary structure of atsRNA is predicted at 37°C by MFOLD program. Bacterial cells containing empty vector pRI were used as the control.