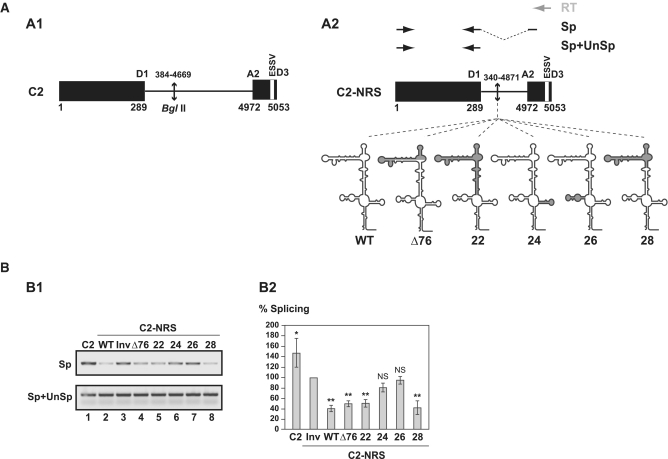
Figure 3.
Comparison of splicing inhibition properties of NRS WT and Δ76, 22, 24, 26 and 28 NRS variants in the C2 pre-mRNA context. (A) Schematic representation of the HIV C2 pre-mRNA (A1) (31) and C2-NRS heterologous pre-mRNAs (A2). Black boxes and thin lines represent exons and intron, respectively. Numbering of C2 RNA is according to (49). End-joining of the two HIV-1 sequences in the C2 intron is depicted by an arrow. The D1 5′-ss and A2 HIV-1 3′-ss as well as the ESSV regulatory element are shown (43). Heterologous C2-NRS constructs, created by insertion of the WT and variant Δ76, 22, 24, 26 and 28 NRSs in the intron, are schematically represented below. Segments deleted in the variant NRS elements are in grey. The RT (in grey), and PCR (in black) primers used for evaluation of the yields of C2 and C2-NRS pre-mRNAs splicing efficiencies are represented by arrows above the C2-NRS construct. Yields of spliced RNAs (Sp) and of spliced plus unspliced products (Sp+UnSp) were analysed by PCR amplifications using the pairs of primers marked by Sp and Sp+UnSp, respectively. (B) RT–PCR analysis of in vitro splicing products of the C2 and heterologous C2-NRS pre-mRNAs. In vitro splicing assays were performed as described in ‘Materials and Methods’ section. Experiments were repeated three times using different batches of RNA (Supplementary Figure S5A). The relative splicing efficiencies of the various constructs were estimated by semi-quantitative RT-PCR performed in triplicate (Supplementary Figure S5A). A representative example of cDNA fractionation obtained for each of the tested pre-mRNAs is shown (B1). Identities of RT–PCR products spliced (Sp) or spliced plus unspliced (Sp+UnSp) products are indicated. The identity of the pre-mRNA used in the assay is given above the lane. The Sp/Sp+UnSp pre-mRNA ratio obtained for the C2-NRS Inv construct was taken as the reference (100%). The Sp/Sp+UnSp ratios established for the other constructs are given as a percentage of this reference. The mean values of the Sp/Sp+UnSp percentages determined for three independent splicing reaction and their standard deviations are given (B2). *P < 0.05, **P < 0.01, NS: not significant.