Figure 1.
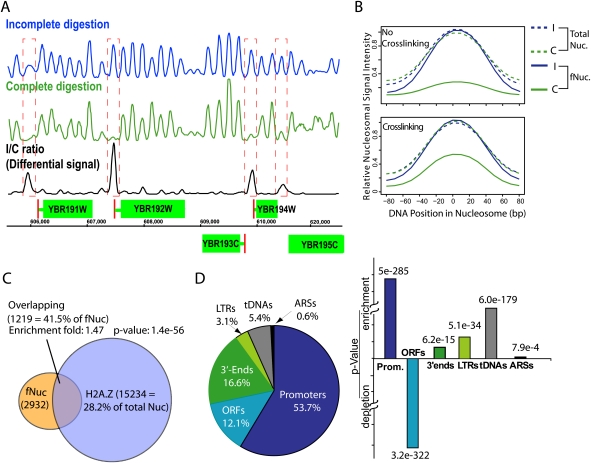
Fragile nucleosomes are localized to the functional regions of the chromatin. (A) Nucleosome occupancy of a region in chromosome II. Plots of MNase-protected DNA sequences illustrate the relative abundance (peak heights) of nucleosomes at particular positions (DNA coordinates, x- axis). (Green boxes) ORFs; (red lines) TSSs. Incomplete/complete differential nucleosome occupancies (fragile nucleosome occupancies; black) are peaks that are missing or significantly reduced in the complete digestion map (green) compared with the incomplete digestion map (blue). (B) Nucleosome fragility is partially suppressed by chemical cross-linking. Panels show the average nucleosomal signal intensity of all nucleosomes (Total Nuc) or the identified 3341 fragile nucleosomes with or without chromatin cross-linking of the chromatin prior to MNase digestion in the incomplete digestion (I, blue) and the complete digestion (C, green) maps, respectively. (C) Partial overlapping between fragile nucleosomes and H2A.Z-nucleosomes (Albert et al. 2007). Enrichment of fragile nucleosomes in H2A.Z nucleosomes is as labeled. (D) Fragile nucleosomes are enriched in chromatin functional regions. (Left) Distributions of fragile nucleosomes in the functional regions. (Right) P-values of fragile nucleosome enrichment or depletion at the labeled functional regions.