FIGURE 3.
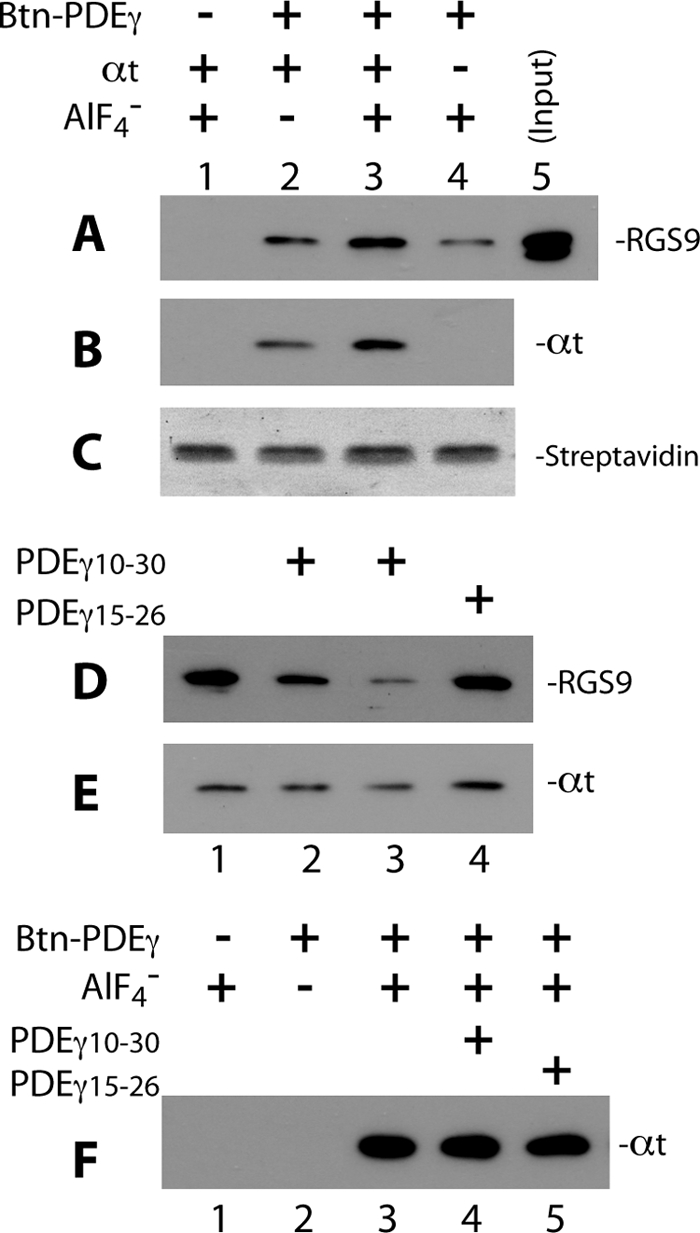
PDEγ N-terminal peptide PDEγ(10–30) disrupts Btn-PDEγ-mediated pulldown of RGS9·β5. Pulldown of RGS9·β5 and αt in the GAP complex was performed using Btn-PDEγ and streptavidin beads. To pulldown RGS9·β5 efficiently, Btn-PDEγ and αt were used in molar excess over the RGS9·β5 heterodimer (molar ratio 25:2.5:1). In each reaction, 2.5 μm Btn-PDEγ was first incubated with RGS9·β5 and αt-GDP on ice for 20 min in Buffer A, the reaction was then mixed with 0.5 μl of streptavidin beads and rotated at 4° C for 20–30 min. 10 mm NaF and 30 μm AlCl3 were added for the plus AlF4− conditions. At the end of incubation, the beads were washed twice with 100 μl of Buffer A. Proteins pulled down on the beads were analyzed by Western blotting. Each blot represents 2–4 similar experiments. A and B, specificity of the pulldown of RGS9 (A) and αt (B), as detected on the same blot, is indicated by lack of signal in the absence of Btn-PDEγ (lane 1) or low signal in the absence of AlF4− (lane 2). Western blotting was first performed with the mouse monoclonal anti-His6 antibody to visualize RGS9, and a secondary probing of αt was done with the rabbit polyclonal anti-αt antibody. C, Amido Black-stained streptavidin bands on the PVDF membrane indicate equal volume of beads used in each reaction. D and E, PDEγ(10–30) but not PDEγ(15–26) reduced RGS9 (D) and αt (E) pulldown. Btn-PDEγ, RGS9·β5, and αt-GDP-AlF4− were present in all conditions. Lane 1 represents exactly the same conditions as those of lane 3 in A and B. PDEγ(10–30) in a molar excess of 150 × (lane 2) or 1,500 × (lane 3) over PDEγ, or PDEγ(15–26)of 150 × (lane 4) was preincubated with RGS9·β5 and αt for 20 min on ice prior to addition of Btn-PDEγ. F, in the absence of RGS9·β5, PDEγ(10–30) (lane 4) or PDEγ(15–26) (lane 5) did not affect αt pulldown by Btn-PDEγ (compared with lane 3). Experiments were conducted under the same conditions as in A–E, except that RGS9·β5 was not added. The −Btn-PDEγ and −AlF4− controls are shown in lanes 1 and 2, respectively.
