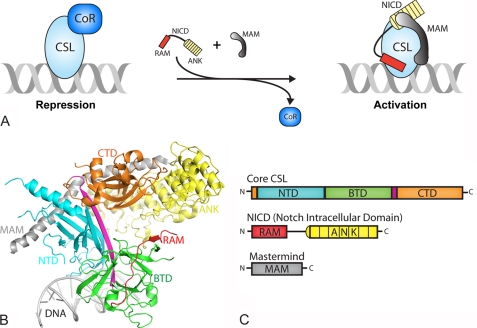
FIGURE 1.
Overview of Notch transcription complexes. A, current model in the field for how transcription is regulated in the Notch pathway. In the absence of a Notch signal, the nuclear effector of the pathway, CSL, specifically binds DNA elements within the promoter and/or enhancer regions of target genes and interacts with transcriptional corepressor proteins in order to repress transcription. Upon pathway activation, the Notch receptor is cleaved at the plasma membrane and its intracellular domain translocates to the nucleus where it interacts with CSL through its RAM and ANK domains. NICD is thought to displace or outcompete corepressors for CSL binding. The CSL-NICD binary complex recruits the transcriptional coactivator MAM, which forms the transcriptionally active CSL-NICD-MAM ternary complex, a necessary step for up-regulating transcription from Notch target genes. B, X-ray structure of the CSL-NICD-MAM ternary complex bound to a cognate DNA (30, 33). CSL is composed of three domains: NTD, BTD, and CTD, which are colored cyan, green, and orange, respectively. A β-strand that makes hydrogen-bonding interactions with all three domains, integrating the three domains into one overall fold, is colored magenta. The NTD and BTD interact with the DNA (gray). NICD interacts with the BTD and CTD through its RAM (red) and ANK (yellow) domains, respectively. MAM, colored gray, forms a bent elongated helix that binds a continuous groove formed by the CTD-ANK interface and the NTD of CSL. C, domain schematics for CSL, NICD, and MAM, coloring is the same as in B.