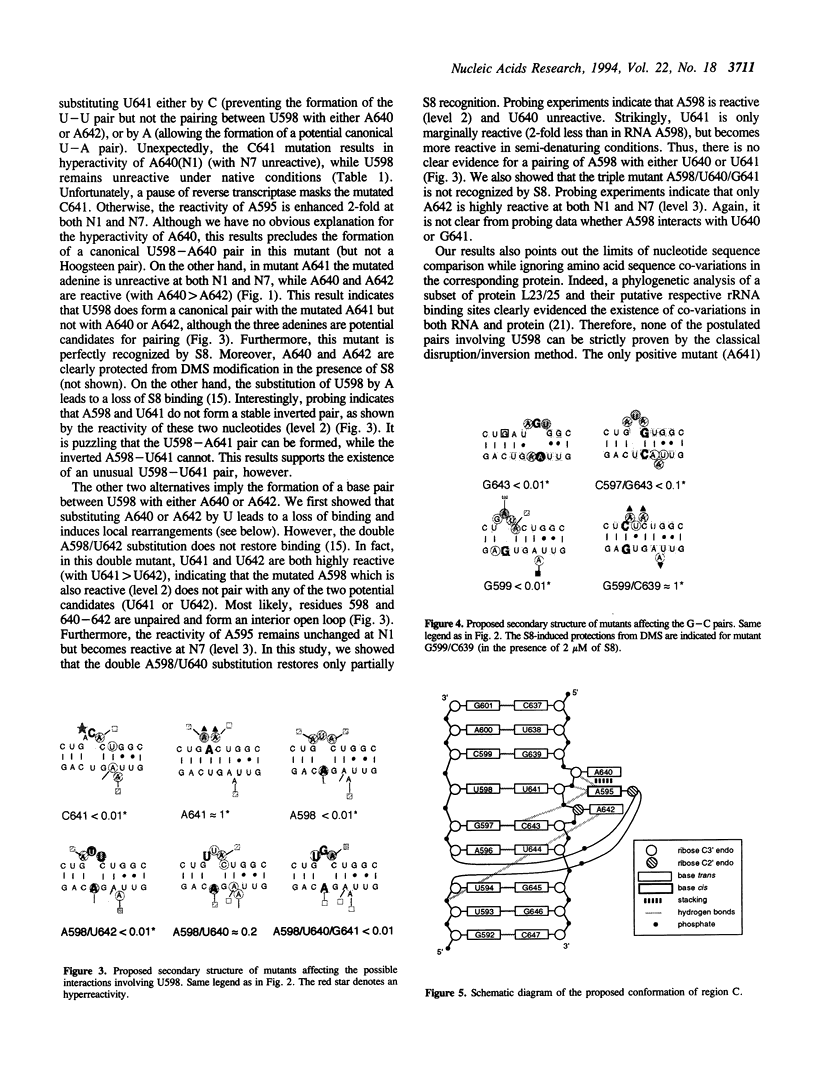
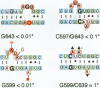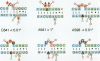Abstract
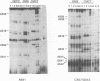
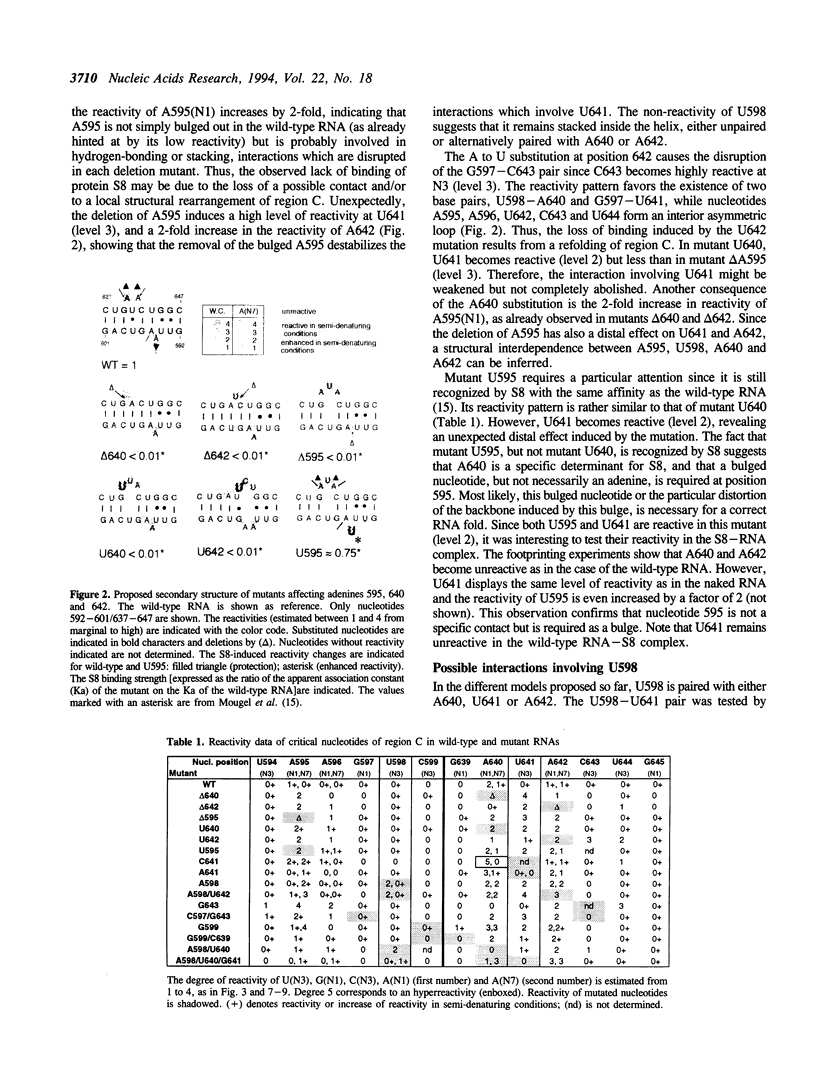
Ribosomal protein S8 specifically recognizes a helical and irregular region of 16S rRNA that is highly evolutionary constrained. Despite its restricted size, the precise conformation of this region remains a question of debate. Here, we used chemical probing to analyze the structural consequences of mutations in this RNA region. These data, combined with computer modelling and previously published data on protein binding were used to investigate the conformation of the RNA binding site. The experimental data confirm the model in which adenines A595, A640 and A642 bulge out in the deep groove. In addition to the already proposed non canonical U598-U641 interaction, the structure is stabilized by stacking interactions (between A595 and A640) and an array of hydrogen bonds involving bases and the sugar phosphate backbone. Mutations that alter the ability to form these interdependent interactions result in a local destabilization or reorganization. The specificity of recognition by protein S8 is provided by the irregular and distorted backbone and the two bulged adenines 640 and 642 in the deep groove. The third adenine (A595) is not a direct recognition site but must adopt a bulged position. The U598-U641 pair should not be directly in contact with the protein.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cachia C., Flamion P. J., Schreiber J. P. Fast preparative separation of 'native' core E coli 30S ribosomal proteins. Biochimie. 1991 May;73(5):607–610. doi: 10.1016/0300-9084(91)90029-z. [DOI] [PubMed] [Google Scholar]
- Cerretti D. P., Mattheakis L. C., Kearney K. R., Vu L., Nomura M. Translational regulation of the spc operon in Escherichia coli. Identification and structural analysis of the target site for S8 repressor protein. J Mol Biol. 1988 Nov 20;204(2):309–329. doi: 10.1016/0022-2836(88)90578-5. [DOI] [PubMed] [Google Scholar]
- Dean D., Yates J. L., Nomura M. Escherichia coli ribosomal protein S8 feedback regulates part of spc operon. Nature. 1981 Jan 1;289(5793):89–91. doi: 10.1038/289089a0. [DOI] [PubMed] [Google Scholar]
- Gregory R. J., Cahill P. B., Thurlow D. L., Zimmermann R. A. Interaction of Escherichia coli ribosomal protein S8 with its binding sites in ribosomal RNA and messenger RNA. J Mol Biol. 1988 Nov 20;204(2):295–307. doi: 10.1016/0022-2836(88)90577-3. [DOI] [PubMed] [Google Scholar]
- Gregory R. J., Zeller M. L., Thurlow D. L., Gourse R. L., Stark M. J., Dahlberg A. E., Zimmermann R. A. Interaction of ribosomal proteins S6, S8, S15 and S18 with the central domain of 16 S ribosomal RNA from Escherichia coli. J Mol Biol. 1984 Sep 15;178(2):287–302. doi: 10.1016/0022-2836(84)90145-1. [DOI] [PubMed] [Google Scholar]
- Gutell R. R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures. Nucleic Acids Res. 1993 Jul 1;21(13):3051–3054. doi: 10.1093/nar/21.13.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held W. A., Ballou B., Mizushima S., Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974 May 25;249(10):3103–3111. [PubMed] [Google Scholar]
- Metzenberg S., Joblet C., Verspieren P., Agabian N. Ribosomal protein L25 from Trypanosoma brucei: phylogeny and molecular co-evolution of an rRNA-binding protein and its rRNA binding site. Nucleic Acids Res. 1993 Oct 25;21(21):4936–4940. doi: 10.1093/nar/21.21.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P. B. On the modus operandi of the ribosome. Cold Spring Harb Symp Quant Biol. 1987;52:721–728. doi: 10.1101/sqb.1987.052.01.081. [DOI] [PubMed] [Google Scholar]
- Mougel M., Allmang C., Eyermann F., Cachia C., Ehresmann B., Ehresmann C. Minimal 16S rRNA binding site and role of conserved nucleotides in Escherichia coli ribosomal protein S8 recognition. Eur J Biochem. 1993 Aug 1;215(3):787–792. doi: 10.1111/j.1432-1033.1993.tb18093.x. [DOI] [PubMed] [Google Scholar]
- Mougel M., Ehresmann B., Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16S rRNA: kinetic and thermodynamic characterization. Biochemistry. 1986 May 20;25(10):2756–2765. doi: 10.1021/bi00358a003. [DOI] [PubMed] [Google Scholar]
- Mougel M., Eyermann F., Westhof E., Romby P., Expert-Bezançon A., Ebel J. P., Ehresmann B., Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16 S rRNA. A model for the interaction and the tertiary structure of the RNA binding site. J Mol Biol. 1987 Nov 5;198(1):91–107. doi: 10.1016/0022-2836(87)90460-8. [DOI] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., De Rijk P., Chapelle S., De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993 Jul 1;21(13):3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson P., Changchien L. M., Craven G. R., Noller H. F. Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16 S ribosomal RNA. J Mol Biol. 1988 Mar 20;200(2):301–308. doi: 10.1016/0022-2836(88)90242-2. [DOI] [PubMed] [Google Scholar]
- Ungewickell E., Garrett R., Ehresmann C., Stiegler P., Fellner P. An investigation of the 16-S RNA binding sites of ribosomal proteins S4, S8, S15, and S20 FROM Escherichia coli. Eur J Biochem. 1975 Feb 3;51(1):165–180. doi: 10.1111/j.1432-1033.1975.tb03917.x. [DOI] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- Wower I., Brimacombe R. The localization of multiple sites on 16S RNA which are cross-linked to proteins S7 and S8 in Escherichia coli 30S ribosomal subunits by treatment with 2-iminothiolane. Nucleic Acids Res. 1983 Mar 11;11(5):1419–1437. doi: 10.1093/nar/11.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. L., Arfsten A. E., Nomura M. In vitro expression of Escherichia coli ribosomal protein genes: autogenous inhibition of translation. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1837–1841. doi: 10.1073/pnas.77.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R. A., Mackie G. A., Muto A., Garrett R. A., Ungewickell E., Ehresmann C., Stiegler P., Ebel J. P., Fellner P. Location and characteristics of ribosomal protein binding sites in the 16S RNA of Escherichia coli. Nucleic Acids Res. 1975 Feb;2(2):279–302. doi: 10.1093/nar/2.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R. A., Singh-Bergmann K. Binding sites for ribosomal proteins S8 and S15 in the 16 S RNA of Escherichia coli. Biochim Biophys Acta. 1979 Jul 26;563(2):422–431. doi: 10.1016/0005-2787(79)90061-3. [DOI] [PubMed] [Google Scholar]
- van den Hoogen Y. T., Treurniet S. J., Roelen H. C., de Vroom E., van der Marel G. A., van Boom J. H., Altona C. Conformational analysis of the tetranucleotides m6(2)A-m6(2)A-U-m6(2)A(m6(2)A = N6-dimethyladenosine) and U-m6(2)A-U-m6(2)A and of the hybrid dA-r(U-A). A one- and two-dimensional NMR study. Eur J Biochem. 1988 Jan 15;171(1-2):155–162. doi: 10.1111/j.1432-1033.1988.tb13771.x. [DOI] [PubMed] [Google Scholar]