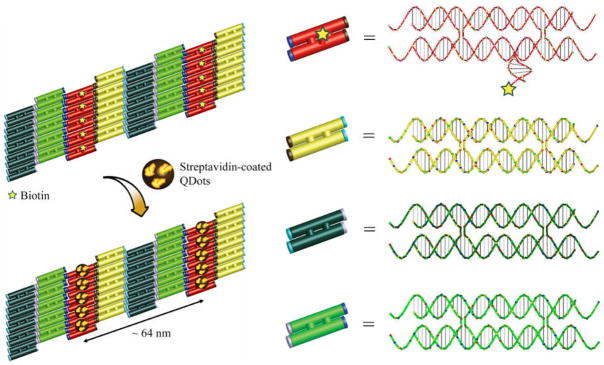
Organizing nanoparticles (NPs) into rationally designed ensemble structures is of great scientific interest because architecturally defined collective properties from multiple NPs could lead to applications such as photonic antennas and controlled plasmonic interactions.[1] Recently, structural DNA nanotechnology has opened a new avenue for directed self-assembly of NPs [2] and other molecular species [3] into patterned nanostructures, taking advantage of the exciting progress in design and construction of artificial nanostructures with complex geometry or patterns via DNA self-assembly.[4] Among these, success of using DNA tile based nanostructure to organize NPs has only been limited to metallic gold NPs. To our knowledge, there has been no report demonstrating DNA tile directed self-assembly of semiconducting NPs (QDs) into rationally designed architectures, partly may be due to the significantly different surface properties of QDs and gold NPs. The difficulty of making QDs compatible to DNA tile based nanostructure has prohibited many interesting studies of multi-component NP photonic systems, e.g. distance dependent plasmonic quenching or enhancement between metallic NPs and QDs. Herein, we worked out a strategy to use two-dimensional DNA tile arrays to direct the assembly of streptavidin conjugated CdSe/ZnS core/shell QDs into well-defined periodic patterns. We anticipate that this first example of DNA tile based QD assembly would pave the way for controlling more sophisticated nanopatterns of QDs and beyond.
The strategy and schematic process of the DNA tile directed QD assembly is illustrated in Figure 1. We used a set of four double crossover (DX) molecules, named the ABCD tile system,[5] as scaffolds for QD assembly. Each different DX tile (DX-A, DX-B, DX-C and DX-D) is shown by a different color in Fig. 1. The ‘A’ tile contains a short DNA stem protruding out of the tile plane that carries a biotin group (illustrated as yellow star) at the end (see supporting information for sequence information). Upon self-assembly, the four-tile system gives 2D arrays displaying parallel lines of biotin groups, with a periodic distance between two neighboring biotin lines ~ 64 nm, and a distance between two biotins within the line about 4–5 nm. After adding streptavidin coated CdSe/ZnS QDs (Invitrogen, QdotR 545 ITK streptavidin conjugate, or STV-QD) to the DNA array, streptavidin molecules (illustrated as yellow blocks) specifically bind to the biotin groups so that the QDs (illustrated as black ball) will be organized onto the DX tile arrays (see supporting information for detailed methods).
Figure 1.
Schematic showing the process of DNA tile directed self-assembly of QD arrays.
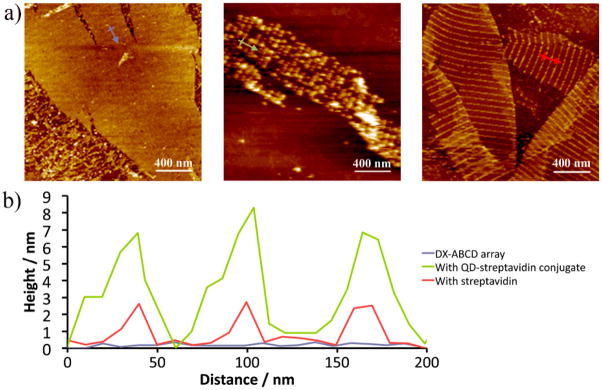
AFM images and cross-section profiles shown in Figure 2 clearly demonstrate that STV-QDs bind specifically to the DX array and get organized into periodical stripes of QD arrays with the designed distances between the parallel lines. Due to the short stem on A tile carrying the biotin group, the patterned 2D array of the ABCD tile system alone (left image and blue trace) show a small height change across the line of the biotin groups, ~ 0.5 nm. When STV-QDs bind to the DX array (middle image and green trace), the average height across the biotin sites increases to ~ 9 nm. A control experiment by adding streptavidin to the same type of biotin modified DX arrays shows a height change of only ~ 2 nm (right image and red trace). The height change resulting from the STV-QD binding is significantly higher than that of streptavidin binding to the array, the line widths are also much wider, owning to much larger sizes of the STV-QDs (diameter estimated ~ 10 nm including the surface polymer and protein coating), compared to that of the protein molecules alone ~ 2–3 nm.
Figure 2.
a) From left to right: AFM images of DX-ABCD array alone with each A tile bearing a biotin; the biotinylated DX-ABCD array incubated with STV-QD conjugate; and same array incubated with streptavidin only; b) Cross section analysis of the AFM images. Each trace corresponds to the same colored arrow in a).
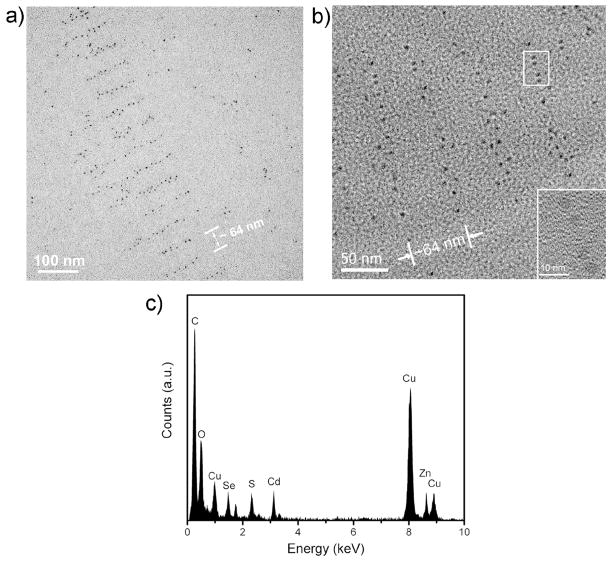
TEM imaging further reveals the patterning of QD arrays templated by the DX tiles. TEM images shown in Figure 3 represent the periodic alignment of QDs into parallel stripes with measured periodicity of ~ 64 nm between the lines, matched well with the designed parameters. The diameter of each individual QD particle is measured ~ 4 nm, corresponds to the size of CdSe/ZnS QDs with green emission. The protein and polymer coating on the surface of the QDs are not visible due to low electron density thus low contrast in the TEM image. It is notable the QDs within each stripe sometimes slightly shift out of the line, which may be due to the orientational flexibility of the short protruding DNA stems bearing the biotin group and the tendency of neighboring QDs to avoid steric crowdedness. The center to center distance between QDs within the same line measures from 7 to 15 nm, which is larger than the distance between neighboring biotin groups, 4–5 nm in the DX array. This can be explained by the large size of the protein coated QDs causing steric hindrance within the line. In addition, the multiple streptavidin molecules conjugated on each QD and the multivalency of streptavidin-biotin binding may also contribute to this effect. It is possible that multiple streptavidin molecules on a single QD may obtain orientations that allow for binding of two or three neighboring biotin groups in the same line. Taking the size of the QD with the protein and polymer coating on surface into account, this 7–15 nm distance is close to the highest possible packing density of the protein coated QDs in the line. More TEM and AFM images of larger sample areas are also included in the supplemental materials, demonstrating the fidelity of the successful assembly of the STV-QDs using the DNA tile scaffolds.
Figure 3.
a) TEM images of the periodic patterns of the organized QD arrays. b) High resolution TEM images with an insert in the right corner reveal the crystalline structure of the QDs. c) EDX spectrum verifies the composition of the CdSe/ZnS QDs.
High resolution TEM images (insert in Fig. 3b) clearly reveals the crystalline structure of the QDs and the energy dispersive X-ray (EDX) data supports that the NPs in the image are composed of CdSe/ZnS, as shown in Figure 3c.
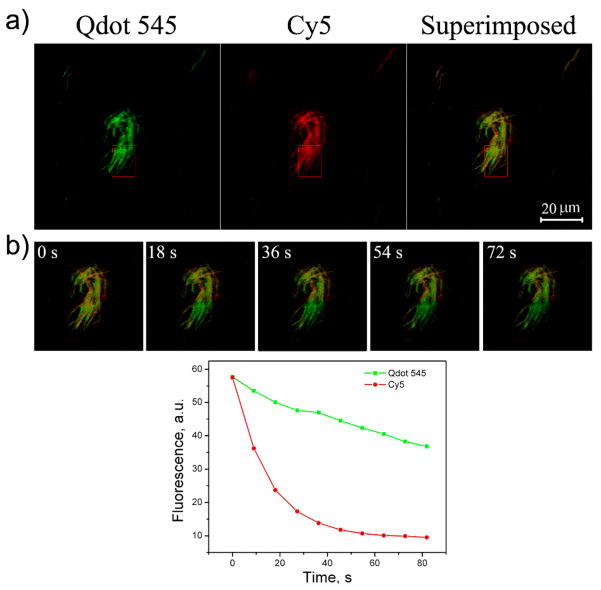
In addition to AFM and TEM imaging, we further used laser fluorescence imaging and photo-bleaching experiments to demonstrate that the QDs were assembled onto the DX array. In this experiment, a DNA strand in the B tile of the DX array was modified with an organic fluorophore with red emission, Cy5 (λem = 648 nm). The DX arrays carrying both Cy5 and STV-QD of green emission (λem = 545 nm) was imaged by fluorescence imaging (Figure 4a), revealing the co-localization of red Cy5 and green QDs on the DNA array (see superimposed fluorescent image in Fig. 4a, rightmost panel). It is well-known that QDs have higher photostability than organic fluorophores. A rectangular shaped region of 11×15 μm2 was selected from the imaged area to be photobleached (Fig. 4b). This region was constantly irradiated by a focused 405 nm laser beam at the power of 0.9 mW for 81 s. Images were taken using the same sequential scanning set-up with 9 s intervals during the photobleaching process. The change of the relative intensity of the red and green fluorescence in the bleached region was also plotted in Figure 4b. It is clear that the organic fluorophore was photobleached with a 90% drop of intensity within 30 s, while the emission intensity of the green QDs still persists after 80 s. This experiment further indicates that QDs are successfully organized onto the DNA tile arrays.
Figure 4.
a) Confocal fluorescent microscope images and b) photobleaching on the DNA arrays. Scale bar: 20 μM.
In summary, we have constructed well-aligned two-dimensional arrays of QDs with controlled periodicity by coupling DNA self-assembly with streptavidin coated QDs. As an elegant bottom-up method, DNA self-assembly has the inherent advantage in generating programmable nanostructures with rationally designed functionality and nanometer precision in addressability. Our work demonstrates the capability to direct QDs into designer nanoarchitectures, this opens up opportunities to construct discrete nanostructures of multi-component NP systems for energy and biosensing applications. It is worthy to point out that both the surface and bioconjugation chemistry of QDs are much more complicated than gold nanoparticles, this led us to optimize many important experimental parameters to achieve successful QD assembly on DNA tile arrays (see supporting information for further comments). We acknowledge that more robust bioconjugation chemistry on QDs to obtain DNA sequence coded QDs (desirable to have discrete number of DNA oligos displayed on the QD surface) is needed for sequence addressable organization of QDs into more sophisticated architectures. Combining QD assembly strategies with previous success of metallic NP organization, we expect that many new properties of the well-controlled multi-component nanophotonic structures will be revealed. Indeed, the organizational power of structural DNA nanotechnology demonstrated so far is only at its horizon.
Supplementary Material
Footnotes
This research was supported by grants from NSF, NIH, AFOSR, ONR and grants from Arizona State University to H.Y. and faculty start-up funds from ASU to Y. L.
References
- 1.Westcott SL, Oldenburg SJ, Lee TR, Halas NJ. Chem Phys Lett. 1999;300:651–655. [Google Scholar]
- 2.a) Le JD, Pinto Y, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Nano Lett. 2004;4:2343–2347. doi: 10.1021/nl0515495. [DOI] [PubMed] [Google Scholar]; b) Zhang J, Liu Y, Ke Y, Yan H. Nano Lett. 2006;6:248–251. doi: 10.1021/nl052210l. [DOI] [PubMed] [Google Scholar]; c) Sharma J, Chhabra R, Liu Y, Ke Y, Yan H. Angew Chem Int Ed. 2006;45:730–735. doi: 10.1002/anie.200503208. [DOI] [PubMed] [Google Scholar]; d) Deng Z, Tian Y, Lee SH, Ribbe AE, Mao C. Angew Chem Int Ed. 2005;117:3648–3651. [Google Scholar]; e) Li H, Park SH, Reif JH, LaBean TH, Yan H. J Am Chem Soc. 2004;126:418–419. doi: 10.1021/ja0383367. [DOI] [PubMed] [Google Scholar]; f) Lee JH, Wernette DP, Yigit MV, Liu J, Wang Z, Lu Y. Angew Chem Int Ed. 2007;46:9006–9010. doi: 10.1002/anie.200702569. [DOI] [PubMed] [Google Scholar]; g) Aldaye F, Sleiman HF. Angew Chem Int Ed. 2006;45:2204–2209. doi: 10.1002/anie.200502481. [DOI] [PubMed] [Google Scholar]; h) Aldaye F, Sleiman HF. J Am Chem Soc. 2007;129:4130–4131. doi: 10.1021/ja070017i. [DOI] [PubMed] [Google Scholar]
- 3.a) Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]; b) Chhabra R, Sharma J, Ke Y, Liu Y, Rinker S, Lindsay S, Yan H. J Am Chem Soc. 2007;129:10304–10305. doi: 10.1021/ja072410u. [DOI] [PubMed] [Google Scholar]; c) Williams BAR, Lund K, Liu Y, Yan H, Chaput JC. Angew Chem Int Ed. 2007;46:3051–3054. doi: 10.1002/anie.200603919. [DOI] [PubMed] [Google Scholar]; d) He Y, Tian Y, Ribbe AE, Mao C. J Am Chem Soc. 2006;128:12664–12665. doi: 10.1021/ja065467+. [DOI] [PubMed] [Google Scholar]
- 4.a) Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]; b) Feldkamp U, Niemeyer CM. Angew Chem Int Ed. 2006;45:1856–1876. doi: 10.1002/anie.200502358. [DOI] [PubMed] [Google Scholar]; c) Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]; d) Gothelf KV, LaBean TH. Org Biomol Chem. 2005;3:4023–4037. doi: 10.1039/b510551j. [DOI] [PubMed] [Google Scholar]; e) Lin C, Liu Y, Yan H. ChemPhysChem. 2006;7:1641–1647. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 5.Liu F, Sha R, Seeman NC. J Am Chem Soc. 1999;121:917–922. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.