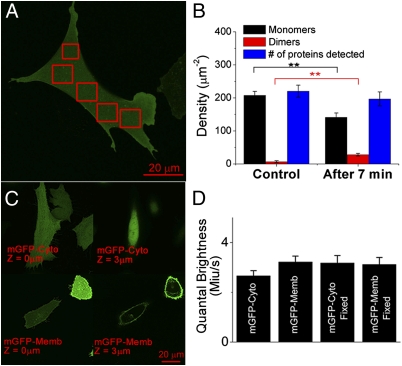
Fig. 3.
Detecting receptor oligomerization in live cells by SpIDA. (A) Image of CHO-k1 cells expressing EGFR-GFP before treatment with EGF. Boxes indicate the ROIs that were analyzed. (B) Shift in the distribution of monomers to dimers after the addition of EGF (20 nM; the experiment was performed at room temperature). The error bars represent the SE taken from five measurements. **P < 0.01 between monomers and dimers for the two cases. No significant difference (P > 0.1) was found for the total number of proteins measured. (C) Images of CHO-k1 cells expressing cytoplasmic mGFPs (mGFP-Cyto) and membrane-targeted mGFP (mGFP-Memb). Two images of the same cell are shown for the two different sample types at two heights in the z stack (0 and 3 μm). (D) Histogram representing the values of the best-fit ε on cells expressing mGFP on the membrane (2D) and cells expressing mGFP in the cytosol (3D). The experiments were performed on both living and fixed cells. For each bar, a minimum N = 20 was sampled. Error bars = SEM. Image size = 1,024 × 1,024 pixels. Pixel size = 0.092 μm. Step size in the z stack = 0.5 μm.