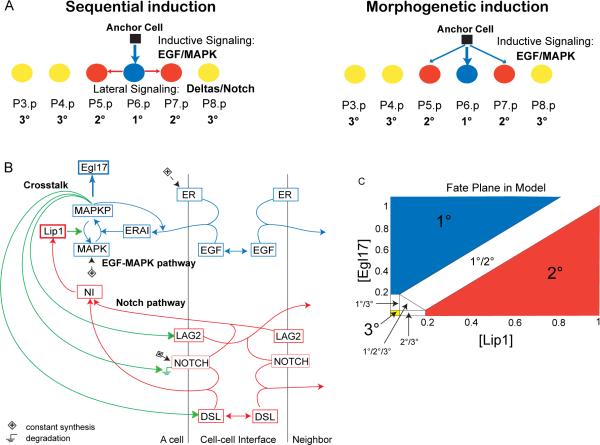
Figure 1. Vulval induction network in C. elegans.
(A) Two verbal models of fate specification in the row of six vulval precursor cells in C. elegans: 1° (blue), 2° (red) and 3° (yellow) fates. (B) Schematic diagram summarizing interactions in the model between EGF (blue) and Notch (red) pathways, and their crosstalk (green). All cells have the same network wiring. Boxes are gene products. ER: EGFR, ERAI: activated EGFR or Ras pathway, MAPKP: phosphorylated MAP kinase, Egl17: 1° cell fate effector, DSL: diffusible Delta, LAG2: membrane-bound Delta, NOTCH: Notch receptor, NI: Notch intracellular domain, Lip1: 2° cell fate effector and MAP kinase phosphatase. The Ground symbols indicate degradation, and (+) constant synthesis. Simple arrows indicate transformation (MAPK to MAPKP and conversely), binding (ligands to receptors) or activation of the downstream node, according to equations in Supplemental Procedures. Double arrows indicate diffusion. (C) Fate plane in the model showing the criteria used to assign cell fates, according to concentrations of Lip1 and Egl17, which are 2° and 1° fate effectors, respectively. Cell fates at the boundaries correspond to (experimentally observed; [11]) intermediate fates. See also Figure S1 and Table S1.