Abstract
Yersinia pestis forms a biofilm in the foregut of its flea vector that promotes transmission by flea bite. As in many bacteria, biofilm formation in Y. pestis is controlled by intracellular levels of the bacterial second messenger c-di-GMP. Two Y. pestis diguanylate cyclase (DGC) enzymes, encoded by hmsT and y3730, and one phosphodiesterase (PDE), encoded by hmsP, have been shown to control biofilm production in vitro via their opposing c-di-GMP synthesis and degradation activities, respectively. In this study, we provide further evidence that hmsT, hmsP, and y3730 are the only three genes involved in c-di-GMP metabolism in Y. pestis and evaluated the two DGCs for their comparative roles in biofilm formation in vitro and in the flea vector. As with HmsT, the DGC activity of Y3730 depended on a catalytic GGDEF domain, but the relative contribution of the two enzymes to the biofilm phenotype was influenced strongly by the environmental niche. Deletion of y3730 had a very minor effect on in vitro biofilm formation, but resulted in greatly reduced biofilm formation in the flea. In contrast, the predominant effect of hmsT was on in vitro biofilm formation. DGC activity was also required for the Hms-independent autoaggregation phenotype of Y. pestis, but was not required for virulence in a mouse model of bubonic plague. Our results confirm that only one PDE (HmsP) and two DGCs (HmsT and Y3730) control c-di-GMP levels in Y. pestis, indicate that hmsT and y3730 are regulated post-transcriptionally to differentially control biofilm formation in vitro and in the flea vector, and identify a second c-di-GMP-regulated phenotype in Y. pestis.
Introduction
Yersinia pestis, the cause of plague, is a Gram-negative bacterium that is transmitted to mammals by infected fleas. It evolved from the enteric pathogen Yersinia pseudotuberculosis within the past 20,000 years [1]. During its life cycle, Y. pestis colonizes the flea midgut and can form a biofilm in the proventricular valve in the foregut. Growth and consolidation of the biofilm within the proventriculus interferes with or completely blocks normal blood feeding, resulting in regurgitation of bacteria and transmission. Fleas with a blocked proventriculus make prolonged, repeated attempts to feed, enhancing the transmission of the bacteria. Unblocked fleas are also capable of spreading disease by early-phase transmission during the first few days after becoming infected [2], [3], but only blocked or partially blocked fleas can transmit disease after the early phase. Thus, the ability to produce a proventricular biofilm is believed to be crucial for long-term enzootic persistence of Y. pestis [3], [4].
When grown at ≤28°C on agar media containing haemin or Congo red (CR), Y. pestis adsorbs the dye and forms greenish-brown or red ‘pigmented’ colonies, respectively. The pigmentation (Pgm+) phenotype of Y. pestis correlates well, although not perfectly, with biofilm formation [5], [6]. Biofilm formation in the flea and in vitro is characterized by a dense aggregate of bacteria embedded within an extracellular matrix (ECM). Y. pestis biofilm and Pgm phenotypes require the hmsHFRS operon, which is responsible for biosynthesis of the ECM polysaccharide [7]–[9].
As in many other bacteria, ECM production and biofilm development in Y. pestis is controlled by bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP), a soluble molecule that functions as a ubiquitous second messenger in bacteria [5], [6], [10]. c-di-GMP stimulates the biosynthesis of adhesin and ECM components and controls the switch between the free-living planktonic and sedentary biofilm-associated lifestyles of many bacteria [11]. c-di-GMP is synthesized by diguanylate cyclase (DGC) enzymes that contain a GGDEF domain and is hydrolyzed by phosphodiesterase (PDE) enzymes that contain an EAL or HD-GYP domain [12]–[14]. Bioinformatic analyses indicate that GGDEF and EAL/HD-GYP domain genes are often highly abundant in bacterial genomes [15]. This redundancy suggests that intracellular levels of c-di-GMP may reflect the cumulative activities of a complex composite of different GGDEF and EAL/HD-GYP family member pairs.
In this study, we evaluated the role of the ten known or putative DGC- and PDE-encoding genes of Y. pestis in regulation of the biofilm phenotype, and show that the two DGCs of Y. pestis make different, environment-dependent contributions to biofilm formation. The DGC encoded by the Y. pestis hmsT gene is sufficient for normal in vitro biofilm formation, whereas the DGC encoded by the y3730 gene has very little effect on in vitro biofilm formation but has the major role in producing proventricular-blocking biofilm in the flea. We also identify autoaggregation as a second c-di-GMP-controlled phenotype in Y. pestis that is unrelated to biofilm exopolysaccharide production.
Results
Effect of Y. pestis GGDEF-, EAL-, and HD-GYP- domain genes on in vitro pigmentation and biofilm phenotypes
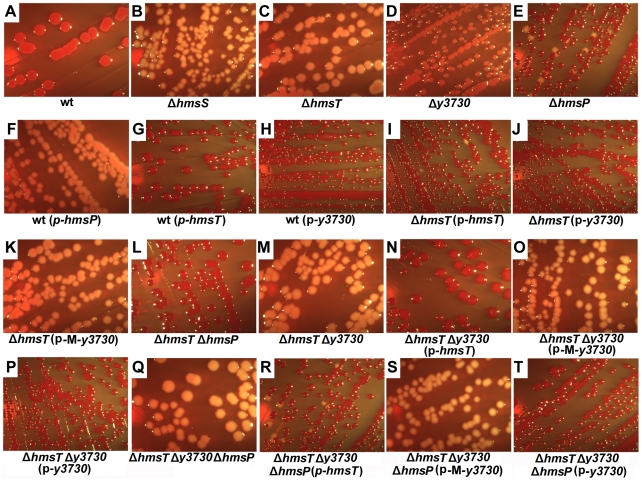
The Y. pestis KIM strain contains ten genes that are predicted to encode GGDEF-domain (Pfam family PF00990), EAL-domain (Pfam family PF00563), or HD-GYP-domain (Pfam family PF0196) proteins (Table 1) [15]–[17]. To determine which of these could potentially participate in proventricular biofilm formation in the flea, we began by evaluating the effect each of these genes had on in vitro biofilm formation. Each gene was individually deleted from Y. pestis KIM6+. The resulting series of mutant strains was first tested for pigmentation phenotype using a formulation of Congo red agar better able to detect relative increases and decreases in pigmentation than the classic Surgalla Congo red agar plates [18]. The plates were incubated at 28°C, a temperature at which the KIM6+ wild-type strain exhibits an intermediate level of pigmentation (Fig. 1A). A known nonpigmented mutant deleted of hmsS was used as a negative control (Fig. 1B). As expected, the hmsT mutant formed nonpigmented colonies and the hmsP mutant formed hyperpigmented colonies that were a much darker red than wild-type colonies (Fig. 1 A, C, E). Deletion of the GGDEF domain gene y3730 resulted in a very subtle decrease in pigmentation (Fig. 1D), but deletion of the other seven genes of interest did not affect the CR phenotype (data not shown).
Table 1. Known and putative DGC and PDE proteins in Y. pestis KIM6+.
| Predicted function | Gene No.a | Gene name | Motif | Y. pstb b homologue | Comparison with Y. pstb homologue |
| DGC | y2559 | - | GGDEF | YPTB1628 | 65% identityc |
| y3730 | - | GGDEF | YPTB0592 | 100% identity | |
| y3756 | hmsT | GGDEF | YPTB0570 | 98% identity | |
| PDE | y1612 | - | ELL | YPTB2605 | 99% identity |
| y2909 | rtn | EAL | YPTB1308 | 93% identityc | |
| y3841 | - | ELL | YPTB3828 | 98% identity | |
| y2472 | - | HI-GYP | YPTB1709 | 30% identityc | |
| DGC and/or PDE | y3389 | - | GGDEF and EAL | YPTB3308 | 92% identityc |
| y3832 | hmsP | SKTEF and EAL | YPTB3836 | 99% identity | |
| Other | y0203 | csrD | LNSDI and EII | YPTB3566 | 99% identity |
Y. pestis KIM annotation number.
Y. pseuodotuberculosis IP32953 annotation number.
Difference due to N-terminal truncation of the predicted Y. pestis protein.
Figure 1. Pigmentation phenotype of Y. pestis strains on LB-Congo red agar.
See text and Table 3 for description of the strains.
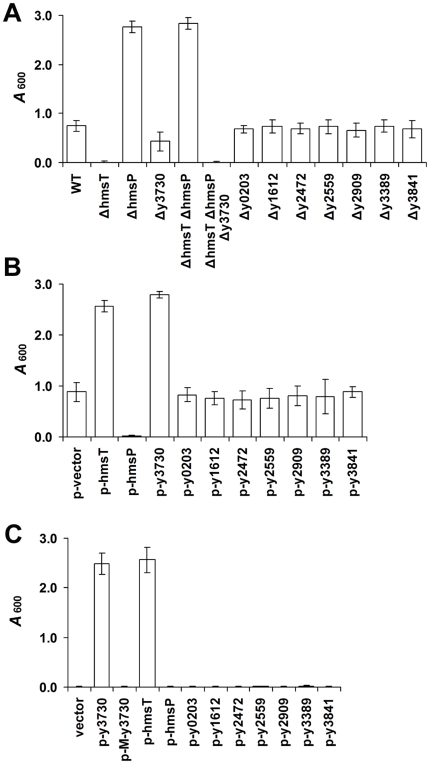
The mutants were further tested for their ability to form biofilm in microtiter plates at room temperature using a crystal violet staining assay (Fig. 2A). Consistent with previous results [6], deletion of hmsT abolished biofilm formation whereas deletion of hmsP resulted in increased biofilm formation. In addition, deletion of y3730 resulted in slightly reduced (not statistically significant) biofilm formation, but deletion of the other seven genes did not affect the ability to produce biofilm in this assay (Fig. 2A).
Figure 2. Effect of Y. pestis GGDEF-, EAL-, and HD-GYP-domain genes on in vitro biofilm formation.
A, B. Relative amounts of adherent biofilm made by Y. pestis KIM6+ parent strain and isogenic derivatives deleted of (A) or overexpressing (B) one of the genes listed in Table 1. C. Quantitation of biofilm made by the hmsT hmsP y3730 triple mutant strain overexpressing one of the genes listed in Table 1, or the mutated y3730 GGAAF allele. The mean and standard deviation of two or more independent experiments are indicated.
To further assess their function, the genes encoding DGC and PDE domains were overexpressed in Y. pestis KIM6+ from a high-copy plasmid vector. As expected, the strain that overexpressed hmsP formed white colonies on the CR plates and the strain that overexpressed hmsT formed hyperpigmented colonies (Fig. 1F, G). The strain that overexpressed y3730 also formed hyperpigmented colonies (Fig. 1H). However, overexpression of any of the other seven genes in Y. pestis KIM6+ did not affect the CR phenotype (data not shown). Complementary results were observed with the microtiter plate biofilm assay, in which increased biofilm was produced only in strains overexpressing hmsT or y3730 (Fig. 2B).
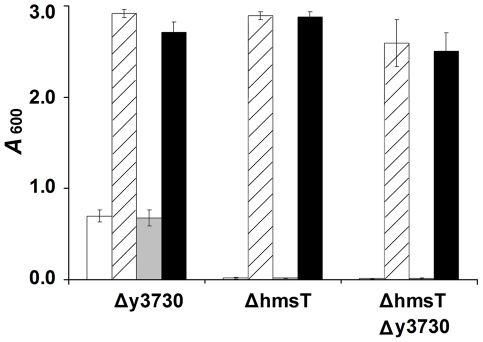
To further confirm that y3730, hmsT, and hmsP are the only c-di-GMP-metabolizing genes involved in biofilm formation, we constructed hmsT y3730 double mutant, hmsT hmsP double mutant, and hmsT hmsP y3730 triple mutant strains of Y. pestis. As predicted, the hmsT hmsP double mutant still formed red colonies on CR plates (Fig. 1L) and strong biofilm in vitro (Fig. 2A), whereas the triple deletion mutant formed white colonies on the CR plate (Fig. 1Q) and no biofilm in vitro (Fig. 2A). Complementation of the hmsT, hmsT y3730 and hmsT hmsP y3730 mutants with either hmsT or y3730 resulted in an equivalent increase in biofilm formation (Fig. 1I-T; Fig. 2C; Fig. 3). However, a plasmid containing a mutated y3730 allele (M- y3730) in which the GGDEF-encoding domain was changed to a GGAAF-encoding domain failed to complement y3730 or hmsT mutation (Fig. 1K,O; Fig. 2C; Fig. 3). Transformation of the triple mutant with high-copy number plasmids containing any of the other eight genes encoding putative DGC and PDE proteins (Table 1) did not change the biofilm-negative and nonpigmented phenotype of the mutant (Fig. 2C and data not shown).
Figure 3. Relative amount of in vitro biofilm made by the Y. pestis y3730, hmsT, and y3730 hmsT mutant strains after transformation with the empty plasmid vector, (white bars) or the plasmid containing wild-type y3730 (hatched bars), mutated y3730 (grey bars), or hmsT (black bars).
The mean and standard deviation of two or more independent experiments are indicated.
Finally, we assayed intracellular levels of c-di-GMP in the Y. pestis hmsT y3730 hmsP triple mutant before and after it was transformed with the p-y3730 plasmid. As predicted, no c-di-GMP was detected in cell lysates of the triple mutant. Synthesis of c-di-GMP was observed when the mutant was transformed with plasmids containing y3730 or hmsT, but not with M-y3730 (Fig. S1).
Differential effect of y3730 and hmsT mutation on Y. pestis biofilm formation in fleas
The preceding results indicated that only the GGDEF domain genes y3730 and hmsT have DGC function that potentially induces proventricular biofilm formation. To investigate the role of y3730 during infection of the flea vector, the Y. pestis KIM6+ parent strain and the y3730 and hmsT mutants were tested for their ability to infect and produce biofilm-dependent proventricular blockage in fleas (Table 2). Infection with Y. pestis KIM6+ resulted in blockage of 38 to 45% of the fleas, consistent with previous results [7]. Although the Y. pestis hmsT mutant forms nonpigmented colonies (Fig. 1C) and little or no biofilm in vitro (Fig. 1C; Fig. 2A, C; Fig. 3), it was still able to block 16 to 20% of infected fleas, a reduction in blockage of about 50% (P<0.0001 by Fisher's exact test). In contrast, although deletion of y3730 had little effect on in vitro pigmentation or biofilm-forming ability (Fig. 1D, Fig. 2A, Fig. 3), proventricular blockage of fleas infected with the y3730 mutant was only 1 to 5%, a rate significantly lower than that of fleas infected with KIM6+ wild-type or hmsT mutant (P<0.0001). The blockage rates produced by the Y. pestis y3730 single mutant and the hmsT y3730 double mutant were not significantly different (P = 0.16), further indicating that y3730 has the predominant role in biofilm formation in the flea. The severe defect in biofilm-dependent proventricular blockage of the y3730 mutant could be restored by complementation with either y3730 or hmsT on a high-copy number plasmid; however, overexpression of the M-y3730 allele lacking the GGDEF domain did not complement the y3730 mutation (Table 2).
Table 2. Infection and blockage of fleas by Y. pestis strains.
| Strain | Y. pestis CFU/fleaa at: | Fleas infected at 28 days | Blockage rate | Fleas(n) | |
| 0 day | 28 days | ||||
| KIM6+ wild type | 8.2×104±5.5×1041.8×105±1.3×105 | 4.8×105±2.4×1056.0×105±3.4×105 | 100%65% | 38%45% | 106113 |
| KIM6+ ΔhmsT | 5.1×104±2.8×1049.2×104±4.9×104 | 3.1×105±1.3×1056.3×105±3.3×105 | 90%80% | 20%16% | 106119 |
| KIM6+ ΔhmsT (p-hmsT) | 1.0×105±7.7×1041.2×105±1.4×105 | 7.7×105±4.5×1055.8×105±4.2×105 | 95%90% | 46%31% | 10590 |
| KIM6+ Δy3730 | 5.8×104±7.3×1044.6×104±4.1×104 | 2.9×105±3.1×1053.2×105±3.4×105 | 70%70% | 1%5% | 88108 |
| KIM6+ Δy3730 (p-y3730) | 4.2×104±4.0×1041.5×105±0.8×105 | 6.4×105±3.7×1057.2×105±2.4×105 | 50%100% | 37%47% | 106106 |
| KIM6+ Δy3730 (p-M-y3730) | 3.3×105±2.2×1054.6×104±2.0×104 | 4.4×105±3.2×1054.3×105±2.7×105 | 95%95% | 8%6% | 108107 |
| KIM6+ Δy3730 (p-hmsT) | 2.7×105±1.8×1051.5×105±0.8×105 | 8.8×105±2.2×1058.5×105±3.0×105 | 95%90% | 42%36% | 101110 |
| KIM6+ ΔhmsTΔy3730 | 1.6×105±1.5×1052.0×104±1.9×104 | 2.7×105±3.9×1053.6×104±4.4×104 | 90%24% | 0%2% | 107109 |
| KIM6+ ΔhmsTΔy3730 (p-y3730) | 6.2×104±5.9×1041.7×104±1.2×104 | 5.0×105±3.6×1055.4×105±3.0×105 | 70%85% | 24%33% | 108111 |
The results of two experiments with each bacterial strain are shown.
Mean±SD.
The infection rate and the mean bacterial load (CFU per flea) at 4 weeks (Table 2) were significantly lower in fleas infected with the Y. pestis hmsT y3730 double mutant compared to fleas infected with wild-type KIM6+ (P<0.05), consistent with previous reports that lack of ability to form biofilm correlates with decreased persistence in the flea [7], [19]. The infection rate and bacterial load in fleas infected with the y3730 mutant was not significantly different than in fleas infected with wild-type or hmsT mutant Y. pestis (Table 2); therefore the highly significant difference in biofilm-dependent blockage produced by the y3730 mutant cannot be accounted for by any decreased ability to produce a chronic infection in the flea gut.
Transcription of y3730 and hmsT is upregulated in the flea
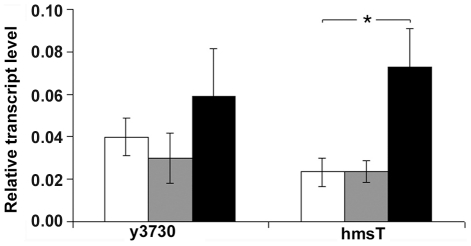
Control of DGC and PDE expression at the transcriptional level in different environments can be important for c-di-GMP regulation in bacteria [10]. We compared transcript levels of hmsT and y3730 in Y. pestis cells isolated from fleas, in vitro biofilms, and planktonic cultures. The expression pattern of both genes was similar in the three different growth conditions, with highest expression detected in the flea (Fig. 4). The relative transcript level of hmsT was even higher than that of y3730 in the flea, but this difference was not statistically significant. Thus, differential transcriptional regulation of hmsT and y3730 does not appear to account for the predominant role of hmsT during in vitro growth or the predominant role of y3730 in the flea.
Figure 4. Expression of Y. pestis y3730 and hmsT in vitro and in the flea.
Relative amounts of y3730 and hmsT mRNA expressed in fleas (black bars), in vitro biofilm cultures (grey bars) and in vitro planktonic cultures (white bars) are shown. The mean and SD of three independent experiments is indicated. *P<0.05 by t-test.
DGC activity is required for the autoaggregation phenotype of Y. pestis
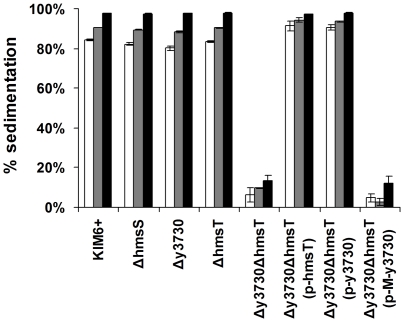
Y. pestis autoaggregates during growth in liquid culture, and this phenotype is not dependent on Hms-dependent ECM or biofilm [20], [21]. In a sedimentation assay to quantitate relative autoaggregation, the Y. pestis hmsS mutant aggregated in culture as rapidly as wild type Y. pestis KIM6+ (Fig. 5), verifying that Hms-dependent ECM is not required for the phenotype. To our surprise, however, disruption of both hmsT and y3730, but not either one alone, resulted in loss of autoaggregation in Y. pestis (Fig. 5). The autoaggregation phenotype of the double mutant could be restored by complementation with either y3730 or hmsT, but not M-y3730. These results suggest that c-di-GMP is involved in the autoaggregation of Y. pestis in an ECM-independent manner.
Figure 5. y3730 and hmsT are required for Y. pestis autoaggregation.
The % sedimentation of bacterial growth in liquid cultures was determined by spectrophotometry after 1, 2, and 12 hours of stasis (white, grey, and black bars, respectively). The mean and SD of two independent experiments performed in duplicate are indicated.
Cyclic-di-GMP signaling pathways are known to control the expression of virulence factors, and autoaggregation is reported to be important for virulence in some bacteria [22]–[24]. Therefore, we also tested the effect of complete loss of Y. pestis c-di-GMP synthetic ability on virulence in a murine model of plague. No significant difference in mortality or time to disease onset was observed in mice injected with the virulent Y. pestis KIM6+ (pCD1-kan) strain or with an isogenic hmsT y3730 (pCD1-kan) double mutant, indicating that loss of DGC activity does not decrease the virulence of Y. pestis for mice (Fig. S2).
Discussion
The biofilm phenotype is important in maintaining flea to mammal transmission cycles of plague, because growth as a biofilm enhances Y. pestis colonization of the flea and transmission by fleabite. Y. pestis also grows as a biofilm in vitro in a temperature-dependent fashion. At temperatures at or below 28°C (corresponding to the ambient temperatures experienced in the flea vector), the ECM required for the pigmentation and biofilm phenotypes is synthesized by the hsmHFRS genes, but at 37°C (corresponding to the mammalian host body temperature), ECM and biofilm are not produced. To date, two mechanisms have been identified that control biofilm development in Y. pestis. As in many bacteria, the second messenger c-di-GMP promotes biofilm development by activating the synthesis of the ECM. Additionally, posttranscriptional regulation by proteolysis of HmsH, HmsR, and HmsT at 37°C is responsible for the lack of biofilm development at high growth temperatures [25].
When we initiated our study, only two enzymes involved in c-di-GMP metabolism, HmsT, a GGDEF-domain diguanylate cyclase, and HmsP, a EAL-domain phosphodiesterase had been characterized in Y. pestis [6], [25]–[27]. A very recent study has identified y3730 as a second DGC gene, also as part of a systematic evaluation of all ten GGDEF-, EAL-, and HD/GYP domain-encoding genes of Y. pestis [28]. This study also demonstrated that HmsT, HmsP, and Y3730 were the only functional c-di-GMP-metabolizing enzymes in Y. pestis, but that y3730 had little effect on the in vitro biofilm phenotype [28]. We have independently confirmed these results, using a somewhat different strategy. We systematically i) deleted and ii) overexpressed all Y. pestis genes that were predicted to encode proteins potentially involved in the metabolism of c-di-GMP and examined the effect on biofilm-related phenotypes. Consistent with the recent study, we also identified y3730 as the only other Y. pestis gene besides hmsT and hmsP that is required for pigmentation and biofilm phenotypes and provide confirmatory data that y3730, like hmsT, encodes a functional GGDEF-domain DGC that synthesizes c-di-GMP.
A striking difference was seen in the relative roles of the two Y. pestis DGCs on biofilm formation in vitro and in the flea. Deletion of hmsT alone virtually eliminated in vitro pigmentation and biofilm phenotypes (Fig. 1C, Fig. 2A, C; Fig. 3), consistent with previous reports [6], [25], [27]. Deletion of y3730, however, resulted in at most a very subtle decrease in in vitro pigmentation and biofilm (Fig. 1D, Fig. 2A), which probably is why y3730 was not discovered in previous random mutagenesis - in vitro screening strategies [27]. In contrast, y3730 deletion had the major effect on in vivo biofilm formation in the flea (Table 2). Differential regulation of hmsT and y3730 transcription does not appear to account for the different in vitro and in vivo phenotypes, because the two genes were transcribed equivalently in both environments, with the expression of both being upregulated in the flea (Fig. 4). One common means by which the enzymatic activity of GGDEF and EAL domain proteins is differentially modulated is via the presence or absence of additional signal input domains [10]. Interestingly, y3730, but not hmsT, encodes a HAMP signaling domain in addition to GGDEF, signal peptide and two widely-spaced transmembrane domains. This pattern is typical of tripartite proteins in which a periplasmic input region responds to an environmental signal that is transduced via the HAMP signal converter to activate a cytoplasmic output domain, in this case GGDEF [29], [30]. Thus, Y3730 may use c-di-GMP as a second messenger to link a specific environmental signal detected only in the flea to the appropriate physiological response– development of a biofilm. If so, the inducing environmental signal presumably is not present or much weaker in vitro, where phenotypes dependent on the DGC activity of Y3730 are not detected unless y3730 is highly overexpressed or hmsP is deleted. This is consistent with the finding that HmsT is responsible for 75–80% of the intracellular c-di-GMP synthesized by culture-grown Y. pestis [28]. Differential stability, localization, or protein-protein interactions of Y3730 and HmsT in the two environments could also contribute to differential activity of the two DGCs in vitro and in the flea. For example, direct interaction among HmsT, HmsP, and other Hms proteins in the inner membrane of Y. pestis has been demonstrated to be important for regulation of biofilm production in vitro [31].
Seven other Y. pestis genes predicted to encode GGDEF-, EAL-, or HD-GYP- domain proteins were also analyzed, but none of them had any effect on pigmentation or biofilm phenotypes (Fig. 2). One of them, y0203, contains degenerate GGDEF and EAL domains and is an ortholog of csrD, which is not involved in c-di-GMP metabolism but controls the degradation of CsrB/CsrC RNAs in E. coli [32]. y1612 and y3841 encode an ELL rather than an EAL domain, and the other four have an N-terminal truncation compared to their Y. pseudotuberculosis orthologs (Table 1). Of these, y2909, y3389, and y2559 have been shown to be pseudogenes in Y. pestis [28].
In summary, our results confirm that only three c-di-GMP metabolizing proteins– two DGCs (HmsT and Y3730) and one PDE (HmsP) are involved in regulation of biofilm formation in Y. pestis, and demonstrate that the role of Y3730 in biofilm production is much greater in the flea than in culture media. We also discovered a second c-di-GMP-dependent phenotype in Y. pestis– autoaggregation. This phenotype is independent of c-di-GMP control of ECM production and may reflect the regulation of non-ECM surface components involved in the biofilm lifestyle. Although c-di-GMP levels are known to regulate the expression of virulence factors and pathogenesis in other bacteria, loss of the two DGCs did not significantly affect pathogenesis in a mouse model of bubonic plague (Fig. S2), suggesting that c-di-GMP does not control the expression of essential virulence factors in Y. pestis. This is consistent with Bobrov et al. [28], who reported that loss of DGC activity did not affect Y. pestis virulence in mouse models of both bubonic and pneumonic plague; and that increased c-di-GMP levels resulted in decreased virulence.
Y. pestis evolved from Y. pseudotuberculosis within the last 20,000 years, while Y. pseudotuberculosis and Y. enterocolitica diverged about 200 million years ago [1], [33], [34]. Y. enterocolitica has 25 genes encoding proteins with GGDEF, EAL, or HD-GYP domains [33], whereas Y. pseudotuberculosis has only ten [34]. Y. enterocolitica and Y. pseudotuberculosis are both food- and water-borne enteric pathogens with similar lifestyles, so it is an interesting question as to why so many DGC and PDE enzymes have been lost in Y. pseudotuberculosis. A complex c-di-GMP signaling pathway network may be beneficial for survival in the environment. For example, Y. pseudotuberculosis can form biofilm in vitro and on the surface of Caenorhabditis elegans nematodes, but not in fleas [35]–[37]. In addition, Y. pseudotuberculosis, unlike Y. pestis, is motile in some environmental conditions, a phenotype that is also often controlled by c-di-GMP. With only three remaining functional DGC and PDE genes, Y. pestis appears to have simplified c-di-GMP signaling pathways even further, with the primary role of enhancing transmissibility by controlling biofilm formation in the flea. It is possible that loss of function of some DGCs and PDEs present in the Y. pseudotuberculosis progenitor might have favored biofilm formation in the flea, and thus mutational loss of these genes may have been positively selected during the evolution of Y. pestis.
Materials and Methods
Bacterial strains and plasmids
The strains and plasmids used are shown in Table 3. The Y. pestis KIM6+ strain was used in this study [38]. A series of mutant KIM6+ strains in which one of each of the ten GGDEF, EAL, or HD-GYP-encoding genes listed in Table 1 was individually deleted and replaced with a chloramphenicol (cat) or kanamycin (kan) resistance gene cassette was made using the λ Red recombination method [39]–[41]. The double hmsT y3730 and triple hmsT y3730 hmsP mutant strains were made similarly by consecutive replacement of the y3730 and hmsP genes with kan and tetracycline resistance genes, respectively, in the ΔhmsT::cat mutant strain.
Table 3. Strains and plasmids used in this study.
| Strain or plasmid | Genotype and/or description | Reference or source |
| Y. pestis KIM6+ strains: | ||
| KIM6+ | wild type | [38] |
| ΔhmsS | ΔhmsS::cat | [40] |
| ΔhmsT | ΔhmsT::cat | This study |
| Δy3730 | Δy3730::kan | This study |
| ΔhmsP | ΔhmsP::kan | This study |
| Δy0203 | Δy0203::kan | This study |
| Δy1612 | Δy1612::kan | This study |
| Δy2472 | Δy2472::kan | This study |
| Δy2559 | Δy2559::kan | This study |
| Δy2909 | Δy2909::kan | This study |
| Δy3389 | Δy3389::kan | This study |
| Δy3841 | Δy3841::kan | This study |
| ΔhmsT Δy3730 | ΔhmsT::cat Δy3730::kan | This study |
| ΔhmsT ΔhmsP | ΔhmsT::cat ΔhmsP::kan | This study |
| ΔhmsT Δy3730 ΔhmsP | ΔhmsT::cat Δy3730::kan ΔhmsP::tet | This study |
| KIM6+ (pCD1-kan) | KIM6+ with pCD1-kan | This study |
| KIM6+ ΔhmsT Δy3730 (pCD1-kan) | KIM6+ ΔhmsT::cat Δy3730::kan with pCD1-kan | This study |
| Plasmids: | ||
| p-hmsT (pCBD26) | hmsT in pCR2.1-TOPO | [41] |
| p-y3730 | y3730 in pUC18 | This study |
| p-hmsP | hmsP in pUC18 | This study |
| p-y0203 | y0203 in pCR2.1-TOPO | This study |
| p-y1612 | y1612 in pCR2.1-TOPO | This study |
| p-y2472 | y2472 in pUC18 | This study |
| p-y2559 | y2559 in pCR2.1-TOPO | This study |
| p-y2909 | y2909 in pCR2.1-TOPO | This study |
| p-y3389 | y3389 in pCR2.1-TOPO | This study |
| p-y3841 | y3841 in pCR2.1-TOPO | This study |
| p-M-y3730 | y3730 with mutated GGDEF domain (D331A,E332A) in pUC18 | This study |
| pCD1-kan | pCD1 virulence plasmid with kan cassette inserted into yadA | This study |
Y. pestis strains overexpressing one of each of the ten GGDEF/EAL/HD-GYP genes (Table 1) were made by first cloning a wild-type copy of each gene, PCR-amplified from KIM6+ using specific primers, into the high-copy number plasmids pUC18 or pCR2.1-TOPO (Invitrogen). These plasmids were then individually transformed into the KIM6+ parent strain by electroporation. A mutated version of the y3730 gene in which the GGDEF-encoding domain was replaced by a GGAAF-encoding domain (resulting in Y3730 D331A, E332A) was prepared by site-specific mutagenesis of the pUC18::y3730 plasmid (p-y3730) using mutagenic primers [42]. The resulting plasmid (p-M-y3730) was used to transform KIM6+. For virulence tests, the KIM6+ and Δy3730 ΔhmsT strains were transformed with the pCD1 virulence plasmid from Y. pestis KIM5 [43] containing a kan gene inserted into the yadA pseudogene. Oligonucleotide primers used for construction of the strains and plasmids are listed in supplemental Table S1. All strains were verified by PCR, DNA sequencing, or plasmid complementation, as appropriate.
Congo red (CR) pigmentation phenotype assays
The strains were streaked on LB agar (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 1.5% agar) supplemented with 0.01% Congo red and colonies were observed visually for pigmentation phenotype (adsorption of the Congo red dye) after growth for two days at 28°C.
Microtiter plate biofilm assays
Bacteria were grown in LB broth supplemented with 4 mM CaCl2 and 4 mM MgCl2 for 24 h at room temperature and diluted to A 600 0.02 in the same medium. 100 µl aliquots were added to wells of 96-well polystyrene dishes, which were incubated with shaking at 250 rpm for 24 h at room temperature. Media and planktonic cells were removed, the wells were washed four times with water, and the adherent biofilm was stained with 200 µl of 0.01% crystal violet for 15 min. The wells were washed four times with water, bound dye was solubilized with 200 µl of 80% ethanol-20% acetone, and the A 600 was measured. Background absorbance for uninoculated control wells was subtracted. The mean and SD was calculated from three independent experiments with at least three replicates.
Flea infections
Xenopsylla cheopis fleas were fed a single infectious blood meal containing ∼5×108 Y. pestis CFU/ml using an artificial feeding system [7], [44]. A sample of 20 female fleas was collected immediately after the infectious blood meal, placed at −80°C, and subsequently used for CFU plate count determinations of the average infectious dose per flea. An additional group of 88 to 119 fleas (approximately equal numbers of males and females) that took an infectious blood meal was maintained at 21°C for four weeks, during which time the fleas were fed twice-weekly on uninfected mice. Immediately after each of these feedings, all fleas were individually examined under a dissecting microscope to determine how many were blocked by proventricular biofilm [7]. After 4 weeks, a sample of 20 surviving female fleas was collected to determine the infection rate and average CFU per infected flea by plate count. Two independent infection experiments were done with each strain. Flea blockage rate and the infection rate at 4 weeks after infection with the different Y. pestis strains were analyzed by two-tailed Fisher's exact test, and differences in the mean CFU per flea at 4 weeks were analyzed by one-way ANOVA with Dunnett's post-test, using GraphPad Prism software.
Quantitative real time PCR
Total RNA of Y. pestis KIM6+ was isolated from cells isolated from in vitro stationary phase planktonic cultures, in vitro biofilms, and infected fleas by using the RNeasy Mini Kit (Qiagen). For the in vitro samples, overnight Y. pestis cultures were diluted to A 600 0.02 in 50 ml LB broth supplemented with 4 mM CalCl2 and 4 mM MgCl2, and grown 24 h with shaking in 250 ml bottles at room temperature. Planktonic cells in the supernatant and biofilm growth adherent to the walls of the culture vessel were separately collected for RNA purification. Y. pestis was recovered from pooled, dissected midguts of fleas two weeks after infection. Residual DNA was removed from the RNA samples by treatment with rDnase I (Ambion) and absence of DNA was confirmed by PCR. cDNA was synthesized from the RNA and used for quantitative PCR on an ABI Prism 7900 sequence detection system (Taqman, Applied Biosytems). The reaction contained oligonucleotide primers, probes and the Taqman universal PCR master mix (Applied Biosystems). The quantity of mRNA for each experimental gene was normalized relative to the quantity of the reference gene crr (y1485), whose expression level is not affected by in vivo or in vitro growth conditions [45]. The ratio of the normalized mRNA quantity of y3730 and hmsT to crr was calculated from three independent assays. Primers and probe sets used are listed in supplemental material Table S1.
Detection of c-di-GMP
c-di-GMP was detected using high-performance liquid chromatography (HPLC) as described previously [46], [47]. Overnight cultures of the Y. pestis ΔhmsTΔy3730 ΔhmsP strain transformed with different recombinant plasmids or with the empty plasmid vector were diluted 1∶30 into 40 ml of LB medium supplemented with 100 mg/l carbenicillin and grown at 28°C for 8 to10 h with shaking at 250 rpm. Formaldehyde (final concentration of 0.18%) was added to cultures and cells were harvested by centrifugation at 3,000 g for 10 min at 4°C. The pellets were washed with 10 mL of phosphate buffered saline (pH 7) with 0.18% formaldehyde and recentrifuged. The bacterial pellets were resuspended in 1 ml of deionized water and heated at 99°C for 15 min. 2 ml of 95% ethanol were added and the lysate was centrifuged. Supernatants were reextracted with 2 ml of 70% ethanol. Pooled supernatants were evaporated, and pellets were dissolved in 100 µl of 0.15 M triethyl ammonium acetate (TEAA, pH 5.0) and centrifuged 15 min at 16000× g. 20 µL of supernatant was fractionated using an Agilent 1200 Series HPLC (Agilent) with a reverse-phase C18 column (Suplecosil LC-18T, 250×4.6 mm, 5 µm; Supelco). Separations were conducted in 0.15 M TEAA at a 1 mL/min flow rate from 20 µl sample injections, using gradient elution with 0 to 15% acetonitrile with detection at 254 nm. Synthetic c-di-GMP (BIOLOG Life Science Institute) was dissolved in 0.15 M TEAA and used as a standard for peak identification and quantification.
Autoaggregation assays
Y. pestis strains were grown overnight at 28°C with shaking at 250 rpm in 15 ml LB supplemented with appropriate antibiotic. Cultures were vortexed, transferred to 15 ml tubes, and allowed to sit undisturbed at room temperature. 1 ml of culture was removed from the top of the tube at t = 0, 1, 2, and 12 h and the A 600 was determined. To calculate the percentage of sedimentation, the A 600 at different time points was divided by the A 600 of the t = 0 sample.
Mouse infection assays
Y. pestis KIM6+ (pCD1::kan) and KIM6+ ΔhmsTΔy3730 (pCD1::kan) strains were grown overnight without shaking in BHI at 28°C and used to inoculate fresh BHI cultures that were incubated overnight in the same conditions. Bacteria were quantified by direct count and serially diluted in PBS. Female 6 to 8-week-old RML Swiss Webster mice were inoculated subcutaneously with 100 CFU of Y. pestis, verified by plating the inocula on blood agar. Mice were examined three times daily for two weeks and euthanized upon signs of terminal plague [44]. Plague was verified by culture of Y. pestis from triturated suspensions of spleens dissected from euthanized mice. All animal experiments were approved by the Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institutes of Health Animal Care and Use Committee (protocol number 07-44) and were conducted in accordance with all National Institutes of Health guidelines.
Supporting Information
Y. pestis y3730 encodes a c-di-GMP synthesizing diguanylate cyclase (DGC) enzyme whose activity is dependent on the GGDEF domain. A. HPLC profile of a solution of 0.4 nmol synthetic c-di-GMP. B–H. HPLC quantification of intracellular levels of c-di-GMP in the Y. pestis hmsT y3730 hmsP triple mutant transformed with the empty plasmid vector (B, C) or with the plasmid vector containing hmsT (D, E), y3730 (F, G) or the mutated GGAAF allele of y3730 (H). Samples C, E, and G were supplemented with 1 nmol synthetic c-di-GMP. Arrows indicate the c-di-GMP peaks.
(TIF)
Loss of diguanylate cyclase (DGC) activity does not significantly reduce Y. pestis virulence in a mouse model of bubonic plague. Incidence of terminal disease in mice after subcutaneous injection of 100 CFU of Y. pestis KIM6+ (pCD1-kan) (white boxes) or KIM6+ ΔhmsT Δy3730 (pCD1-kan) (black circles) is shown. All ten mice developed terminal plague after injection of the wild type Y. pestis KIM6+, and 9 of 10 mice developed terminal plague after injection of the ΔhmsT Δy3730 double mutant.
(TIF)
Sequences of primers and probes used in this study.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH); and NIH Grant AI057512 (to C.D.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, et al. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, et al. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci U S A. 2006;103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisen RJ, Wilder AP, Bearden SW, Montenieri JA, Gage KL. Early-phase transmission of Yersinia pestis by unblocked Xenopsylla cheopis (Siphonaptera: Pulicidae) is as efficient as transmission by blocked fleas. J Med Entomol. 2007;44:678–682. doi: 10.1603/0022-2585(2007)44[678:etoypb]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Eisen RJ, Gage KL. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet Res. 2009;40:1. doi: 10.1051/vetres:2008039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinnebusch BJ, Erickson DL. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr Top Microbiol Immunol. 2008;322:229–248. doi: 10.1007/978-3-540-75418-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol. 2004;54:75–88. doi: 10.1111/j.1365-2958.2004.04253.x. [DOI] [PubMed] [Google Scholar]
- 7.Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 8.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- 9.Perry RD, Pendrak ML, Schuetze P. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J Bacteriol. 1990;172:5929–5937. doi: 10.1128/jb.172.10.5929-5937.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 11.Jonas K, Melefors O, Romling U. Regulation of c-di-GMP metabolism in biofilms. Future Microbiol. 2009;4:341–358. doi: 10.2217/fmb.09.7. [DOI] [PubMed] [Google Scholar]
- 12.Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci U S A. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 16.Deng W, Burland V, Plunkett G, 3rd, Boutin A, Mayhew GF, et al. Genome sequence of Yersinia pestis KIM. J Bacteriol. 2002;184:4601–4611. doi: 10.1128/JB.184.16.4601-4611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surgalla MJ, Beesley ED. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl Microbiol. 1969;18:834–837. doi: 10.1128/am.18.5.834-837.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vetter SM, Eisen RJ, Schotthoefer AM, Montenieri JA, Holmes JL, et al. Biofilm formation is not required for early-phase transmission of Yersinia pestis. Microbiology. 2010;156:2216–2225. doi: 10.1099/mic.0.037952-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felek S, Muszynski A, Carlson RW, Tsang TM, Hinnebusch BJ, et al. Phosphoglucomutase of Yersinia pestis is Required for Autoaggregation and Polymyxin B Resistance. Infect Immun. 2009;78:1163–1175. doi: 10.1128/IAI.00997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolodziejek AM, Sinclair DJ, Seo KS, Schnider DR, Deobald CF, et al. Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology. 2007;153:2941–2951. doi: 10.1099/mic.0.2006/005694-0. [DOI] [PubMed] [Google Scholar]
- 22.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, et al. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun. 2007;75:3233–3244. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang SL, Taylor RK, Koomey M, Mekalanos JJ. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol Microbiol. 1995;17:1133–1142. doi: 10.1111/j.1365-2958.1995.mmi_17061133.x. [DOI] [PubMed] [Google Scholar]
- 24.Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Perry RD, Bobrov AG, Kirillina O, Jones HA, Pedersen L, et al. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J Bacteriol. 2004;186:1638–1647. doi: 10.1128/JB.186.6.1638-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hare JM, McDonough KA. High-frequency RecA-dependent and -independent mechanisms of Congo red binding mutations in Yersinia pestis. J Bacteriol. 1999;181:4896–4904. doi: 10.1128/jb.181.16.4896-4904.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones HA, Lillard JW, Jr, Perry RD. HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology. 1999;145(Pt 8):2117–2128. doi: 10.1099/13500872-145-8-2117. [DOI] [PubMed] [Google Scholar]
- 28.Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, et al. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol Microbiol. 2010;79:533–551. doi: 10.1111/j.1365-2958.2010.07470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Appleman JA, Chen LL, Stewart V. Probing conservation of HAMP linker structure and signal transduction mechanism through analysis of hybrid sensor kinases. J Bacteriol. 2003;185:4872–4882. doi: 10.1128/JB.185.16.4872-4882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 31.Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol. 2008;10:1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki K, Babitzke P, Kushner SR, Romeo T. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 2006;20:2605–2617. doi: 10.1101/gad.1461606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson NR, Howard S, Wren BW, Holden MT, Crossman L, et al. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2006;2:e206. doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature. 2002;417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- 36.Erickson DL, Jarrett CO, Callison JA, Fischer ER, Hinnebusch BJ. Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J Bacteriol. 2008;190:8163–8170. doi: 10.1128/JB.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshua GW, Karlyshev AV, Smith MP, Isherwood KE, Titball RW, et al. A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology. 2003;149:3221–3229. doi: 10.1099/mic.0.26475-0. [DOI] [PubMed] [Google Scholar]
- 38.Fetherston JD, Schuetze P, Perry RD. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 39.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun YC, Hinnebusch BJ, Darby C. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc Natl Acad Sci U S A. 2008;105:8097–8101. doi: 10.1073/pnas.0803525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun YC, Koumoutsi A, Darby C. The response regulator PhoP negatively regulates Yersinia pseudotuberculosis and Yersinia pestis biofilms. FEMS Microbiol Lett. 2009;290:85–90. doi: 10.1111/j.1574-6968.2008.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li QX, Dowhan W. Studies on the mechanism of formation of the pyruvate prosthetic group of phosphatidylserine decarboxylase from Escherichia coli. J Biol Chem. 1990;265:4111–4115. [PubMed] [Google Scholar]
- 43.Perry RD, Straley SC, Fetherston JD, Rose DJ, Gregor J, et al. DNA sequencing and analysis of the low-Ca2+-response plasmid pCD1 of Yersinia pestis KIM5. Infect Immun. 1998;66:4611–4623. doi: 10.1128/iai.66.10.4611-4623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erickson DL, Jarrett CO, Wren BW, Hinnebusch BJ. Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis. J Bacteriol. 2006;188:1113–1119. doi: 10.1128/JB.188.3.1113-1119.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sebbane F, Lemaitre N, Sturdevant DE, Rebeil R, Virtaneva K, et al. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci U S A. 2006;103:11766–11771. doi: 10.1073/pnas.0601182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simm R, Fetherston JD, Kader A, Romling U, Perry RD. Phenotypic convergence mediated by GGDEF-domain-containing proteins. J Bacteriol. 2005;187:6816–6823. doi: 10.1128/JB.187.19.6816-6823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Y. pestis y3730 encodes a c-di-GMP synthesizing diguanylate cyclase (DGC) enzyme whose activity is dependent on the GGDEF domain. A. HPLC profile of a solution of 0.4 nmol synthetic c-di-GMP. B–H. HPLC quantification of intracellular levels of c-di-GMP in the Y. pestis hmsT y3730 hmsP triple mutant transformed with the empty plasmid vector (B, C) or with the plasmid vector containing hmsT (D, E), y3730 (F, G) or the mutated GGAAF allele of y3730 (H). Samples C, E, and G were supplemented with 1 nmol synthetic c-di-GMP. Arrows indicate the c-di-GMP peaks.
(TIF)
Loss of diguanylate cyclase (DGC) activity does not significantly reduce Y. pestis virulence in a mouse model of bubonic plague. Incidence of terminal disease in mice after subcutaneous injection of 100 CFU of Y. pestis KIM6+ (pCD1-kan) (white boxes) or KIM6+ ΔhmsT Δy3730 (pCD1-kan) (black circles) is shown. All ten mice developed terminal plague after injection of the wild type Y. pestis KIM6+, and 9 of 10 mice developed terminal plague after injection of the ΔhmsT Δy3730 double mutant.
(TIF)
Sequences of primers and probes used in this study.
(DOC)