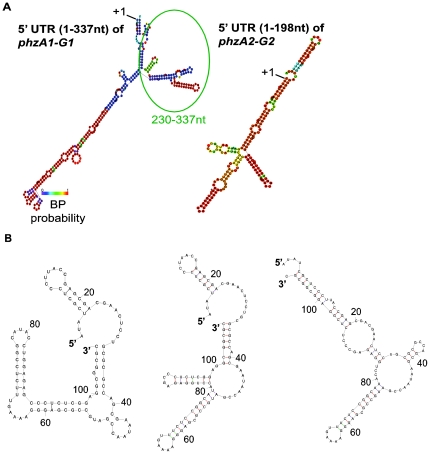
Figure 4. Secondary structures in the 5′-UTRs of two phz gene clusters were predicted by RNA fold.
The predicted secondary structures in 5′-UTR of phzA1-G1 (1–337 nt) and phzA2-G2 (1–198 nt) gene clusters (A). The conserved RNA secondary structures were predicted from the alignment of Pseudomonas sp. M18 and P. aeruginosa PA7. The base pairing probabilities were annotated with colors. Three suboptimal secondary structures were also predicted for a portion of the 5′-UTR of the phzA1-G1 gene cluster in Pseudomonas sp. M18 (B). Gibbs free energies (ΔG) of the three suboptimal local structures from left to right are −39.3 kcal/mol, −38.1 kcal/mol and −36.7 kcal/mol, respectively.