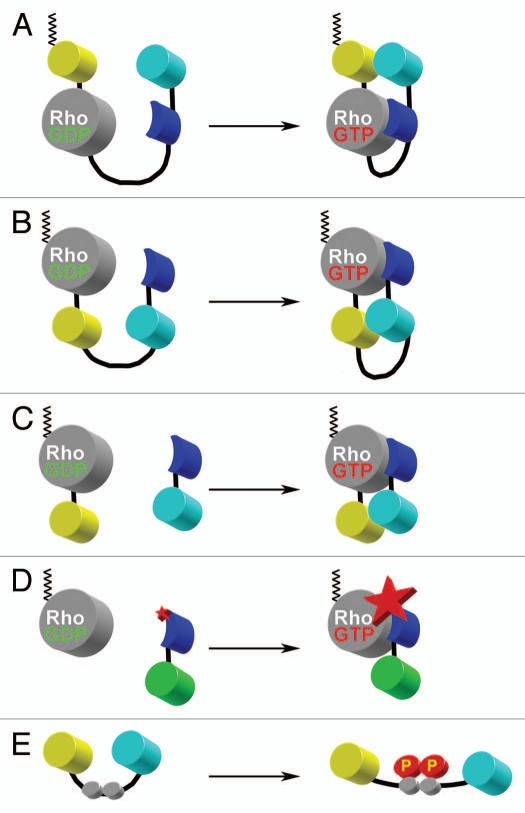
Figure 4.
Schematic representations of various design approaches used in FRET biosensors. (A) Single-chain, genetically-encoded biosensor for Rho family GTPases based on the design of Raichu-sensors for Ras.135 This design places a CAAX box from K-Ras at the C-terminal end of the fluorescent protein to constitutively place the probe into the plasma membrane (depicted in black zig-zag lines). Upon activation of Rho GTPase through GDP-GTP exchange, the binding domain derived from a downstream effector target (blue) binds to the GTPase, changing the relative proximities of the FRET pair of yellow and cyan fluorescent proteins. (B) Single-chain, genetically-encoded biosensor for Rho GTPase.48 This design places the FRET pair of fluorescent proteins within the internal portion of the single-chain construction. This approach maintains the native C-terminus of the Rho GTPase where proper post-translational lipid modification can take place. This feature maintains the interaction between the GTPase and the guanine nucleotide dissociation inhibitor. This allows for the proper shuttling of the biosensor between the cytoplasm (inactive) and the plasma membrane (active), in a manner analogous to the endogenous protein. Upon activation of the GTPase through GDP-GTP exchange, the binding domain (blue) binds to activated Rho, changing the relative orientations between the cyan and yellow fluorescent proteins to affect FRET. (C) Bi-molecular, genetically encoded biosensor for Rho GTPases.37 This design separates the FRET donor and acceptor moieties into two molecules which must be expressed separately. While the quantitative analysis is more cumbersome due to non-equimolar distribution of the FRET donor and acceptor pairs, the total change in dynamic range can be much greater than any single-chain versions of the biosensor. (D) Solvent-sensitive dye-based biosensor for detecting the activity of an endogenous Rho GTPase.47 This approach uses an organic dye that changes the fluorescence emission intensity as a function of the local solvent polarity. By placing the dye within the binding domain (blue) derived from a downstream target protein, the binding of this biosensor to the activated, unlabeled, endogenous Rho GTPase causes the dye to change the fluorescence intensity of emission. By monitoring the ratio of this fluorescence emission intensity modulation over the non-responsive fluorescence (e.g., green fluorescent protein shown in green), one can monitor the effector-binding state of the Rho GTPase. (E) Substrate-based FRET biosensor for the kinase activity. The FRET pair of fluorescent proteins flank the phosphorylation motif of the target upstream kinase (gray circles). Upon phosphorylation, the conformation of the molecule is slightly altered and affects the FRET between the two fluorescent proteins. This approach can also include an appropriate binding site adjacent to the kinase phosphorylation motif for the upstream kinase of interest in order to improve the selectivity (not shown). The main caveat to this approach is that it is not as specific for the “kinase activity” because it simply monitors the balance of the relative upstream kinase and the local phosphatase activities in live-cell situations.