Abstract
This is an open access article. Unrestricted non-commercial use is permitted provided the original work is properly cited.
Aberrant microRNA activity has been reported in many diseases, and studies often find numerous microRNAs concurrently dysregulated. Most target genes have binding sites for multiple microRNAs, and mounting evidence indicates that it is important to consider their combinatorial effect on target gene repression. A recent study associated the coincident loss of expression of six microRNAs with metastatic potential in breast cancer. Here, we used a new computational method, miR-AT!, to investigate combinatorial activity among this group of microRNAs. We found that the set of transcripts having multiple target sites for these microRNAs was significantly enriched with genes involved in cellular processes commonly perturbed in metastatic tumors: cell cycle regulation, cytoskeleton organization, and cell adhesion. Network analysis revealed numerous target genes upstream of cyclin D1 and c-Myc, indicating that the collective loss of the six microRNAs may have a focal effect on these two key regulatory nodes. A number of genes previously implicated in cancer metastasis are among the predicted combinatorial targets, including TGFB1, ARPC3, and RANKL. In summary, our analysis reveals extensive combinatorial interactions that have notable implications for their potential role in breast cancer metastasis and in therapeutic development.
Keywords: microRNA, cancer, metastasis, bioinformatics, computational
Introduction
Involved in the regulation of many cellular processes, microRNAs are short, endogenous oligonucleotides that have been implicated in a wide variety of diseases. The mature form of microRNAs (miRNAs) are approximately 22 nucleotides in length and anneal to complementary sites in the 3′ untranslated region (UTR) of target transcripts as part of an RNA-induced silencing complex (RISC). While microRNAs appear to act through multiple mechanisms, two general modes of action have been identified: transcript degradation and inhibition of protein translation.1–3 The former is associated with near-perfect base complementarity between a miRNA and its target sequence, while non-perfect miRNA-target matches result in inhibition of protein translation. A number of recent reports have shown that non-perfect complementarity can also result in mRNA degradation through poly(A) deadenylation.4,5 Therefore, the effect of microRNA activity may be reflected by changes of either mRNA or protein levels, depending on the microRNA and target transcript involved. There are currently over 700 known miRNAs in the human genome, and each miRNA may regulate dozens to hundreds of target transcripts. Reports have shown numerous miRNAs aberrantly expressed in a variety of diseases, including cancer, and indicate that hundreds and perhaps thousands of transcript targets could potentially be affected in neoplastic tissue. The 3′ UTR in a single messenger RNA may contain binding sites for a number of microRNAs, and a transcript can be concurrently repressed by multiple microRNA species.6 Therefore, the genome-wide complexity of microRNA interactions presents a formidable challenge to understanding cellular regulation, particularly when assessing the impact of multiple dysregulated miRNAs.
Mounting evidence reveals that microRNAs can exert a cooperative effect on target gene repression. Since a gene product may be simultaneously repressed by multiple microRNAs, and a number of microRNAs may be differentially expressed in a given disease condition, it is essential to consider their combinatorial interactions.1,3,7,8 A recent genome-wide investigation demonstrated a significant and positive correlation between the number of microRNA binding sites in a transcript and the mRNA decay rate.9 This additive effect is believed to be due primarily to the combinatorial contribution of multiple microRNAs since only a small fraction of transcripts contain multiple binding sites for any single miRNA. The authors noted that the vast majority of target genes have sites for multiple microRNA species, thus providing a mechanism for cooperative repression of target genes. Another recent study identified combinatorial activity among four microRNAs involved in monocyte differentiation.10 Concurrent over-expression of the four microRNAs resulted in numerous gene expression changes that were not observed with any one of the four individual miRNA transfections, demonstrating cooperative interactions. Two microRNAs in a bicistronic cluster (miR-143 and miR-145) were reported to be repressed in most gastric and colon cancer patients that were tested.11 By over-expressing these microRNAs in a gastric cancer cell line, the authors confirmed an additive effect on target gene repression, and their observations indicate combinatorial activity by the two microRNAs. A separate study demonstrated that while these two microRNAs are nonhomologous, they share numerous common targets involved in actin dynamics and cytoskeletal function, suggesting cooperative regulation of cellular pathways.12 Based on growing evidence of cooperative microRNA activity, it is imperative to consider and investigate combinatorial effects where multiple dysregulated microRNAs are identified in expression profiling studies.
While several computational tools have been developed to address components of this goal, there is a need for freely available software that can perform combinatorial target analysis of microRNAs and functional annotation of the target genes.7,13–16 A recent review on computational methods for microRNA studies called for the development of tools for combinatorial analysis and noted that “it is important to develop novel computational methods that explicitly capture dependencies between individual miRNA targeting and reveal synergistic effects on functionally related genes”.17 Here, we introduce a web-based bioinformatics application, miR-AT! (microRNA Combinatorial Analysis of Targets), which leverages and integrates several existing, high-quality databases to enable combinatorial microRNA target characterization and functional analysis. Among the features of miR-AT! are: the ability to predict combinatorial targets of multiple microRNAs, user specified parameters for minimum number of sites and number of unique microRNAs found in each target, minimum score criteria, integrated functional annotation of predicted targets, and a novel clustering implementation that enables identification of transcripts with similar microRNA target site patterns. In this work we have applied miR-AT! to investigate combinatorial activity associated with a set of microRNAs implicated in breast cancer metastasis.
Tavazoie et al recently identified six microRNAs (hsa-mir-335, 126, 206, 122, 199a-3p, and 489) that were significantly decreased in highly metastatic breast cancer cell derivatives.18 All six of the microRNAs were consistently downregulated in cell derivatives that aggressively metastasized to bone or lung, as compared to the parental cells. The authors focused on this set of miRNAs based on the combined results of microarray profiling and RT-PCR, and subsequently investigated the effect of each of the six microRNAs by individually restoring expression with retroviral transduction. It was determined that individually, miR-335, miR-206, and miR-126 had the greatest effect on metastasis, and restoration of either of these microRNAs resulted in a significant decrease in lung colonization at the end point. Restoration of either of miR-122, miR-199a-3p, or miR-489 also decreased colonization, but at earlier timepoints. The six microRNAs reside on five different chromosomes, and there is no evidence of co-regulation of these microRNAs. The coincident loss of all six microRNAs in each of the highly metastatic cell derivatives suggests that cooperative activity among them may be important in maintaining post-transcriptional regulation of target genes involved in tumor growth and metastasis. We have used miR-AT! to perform computational target analysis of the aggregate set of six miRNAs, considering potential combinatorial activity and multiple microRNA binding sites in predicted target genes.
Methods
miR-AT!
miR-AT! is a web accessible application available at http://mir-at.org. A user guide is provided through the “help” link available at the home page. The application leverages several high quality databases and software applications to provide target prediction and subsequent functional annotation of target genes. Additionally, miR-AT! provides the capability to cluster target genes by the pattern of microRNA binding sites in their 3′ UTR.
From a user-submitted list of miRNA identifiers, the miR-AT! application scans a local implementation of the MicroCosm Targets database, formerly known as miRBase Targets (http://microrna.sanger.ac.uk/targets/v5/).19 The application identifies all transcripts having predicted target sites matching the microRNAs provided in the input set and meeting user-specified score criteria. Target identification in MicroCosm Targets is accomplished using 3′ UTRs of Ensembl transcripts and the miRanda algorithm,20 allowing no more than one mismatch in the critical “seed” region on the 5′ end of the miRNA. Each target site in the database has an associated score and P-value. User selectable parameters in miR-AT! allow the specification of a minimum score required for each target site and a maximum P-value allowed. The user can specify a minimum number of total target sites required for each transcript, and these can be satisfied by any combination of the input microRNAs. Additionally, a minimum number of unique microRNAs represented in each transcript can be specified. miR-AT! currently recognizes miRNA identifiers for human, mouse, and rat. Addition model organisms will be added in the future.
A range of computational methods have been developed for microRNA target site prediction, but few have included consideration of combinatorial effects. Of established methods, perhaps the two most closely aligned with our objective of investigating microRNA combinatorial activity are miRGator and microRNA.org.15,21 Both methods allow for the input of multiple microRNAs, and both report targets having one or more predicted sites associated with the input microRNA set. However, miR-AT! provides additional capabilities essential to investigating combinatorial effects, including the ability to specify a minimum number of target sites per gene, a minimum number of unique microRNAs from the input set that must target each gene, and filtering tools for individual target site scores and P-values. Furthermore, miRGator restricts queries to fewer than five microRNAs, and microRNA.org is limited to searches for targets of one or all of the input microRNAs, not intermediate combinations. Collectively, miR-AT! provides greater flexibility to construct combinatorial queries and enables the selection and filtering of high quality targets based on site number, score, and significance. Additionally, the target score distribution analysis tool available in miR-AT! allows the user to visualize a histogram of scores associated with the target gene set, thus providing assistance in identifying cutoff scores to remove low confidence target predictions.
In addition to the identification of combinatorial target gene sets, an important feature of miR-AT! is the ability to submit the target sets to DAVID for comprehensive functional annotation and pathway analysis. MirGator and microRNA.org do not provide functional annotation capability for combinations of microRNAs. Another unique feature of miR-AT! is the ability to cluster target genes based on the combinatorial pattern of target sites found in each 3′ UTR. This feature identifies groups of genes that have similar target site combinations and thus are predicted to be under similar influence of the input microRNAs. The method also clusters the input microRNAs based on their target site patterns in the identified target gene set. The clustering tool can be used to explore the predicted combinatorial effect of microRNAs on sub-clusters of target genes and also to identify microRNAs having potential cooperative activity as discussed below and exemplified by our findings with miR-122 and miR-206.
In this work we submitted six microRNAs previously identified as repressed in highly metastatic breast cancer cells: hsa-miRs 335, 126, 206, 122, 199a-3p, and 489. We used the synonymous designations of hsa-mir-199a-3p and hsa-mir-122 for 199a* and 122a respectively, the latter having been used in the Tavazoie report.
Target list output
Each target with a miRNA site satisfying the specified selection criteria is included in the output list of transcripts. miR-AT! calculates a cumulative score for each transcript, derived from the sum of all miRNAs meeting the specified selection criteria. The cumulative score provides for the identification of transcripts most likely affected by the combined set of input miRNAs. Multiple bound miRNAs can additively affect messenger RNA translation and/or stability, thus it is crucial to consider the cumulative set of miRNA sites when assessing putative targets. By default, the output list is sorted by the cumulative score. Thus, targets with multiple, high scoring sites will be found towards the top of the list and transcripts with a lower cumulative score will be found towards the bottom. For each target in the output list the NCBI Gene ID, Ensembl gene and transcript identifiers, description, number of target sites, and cumulative score are provided. The NCBI and Ensembl IDs are hyperlinked to the respective databases to provide extensive information for each target transcript. The total number of miRNA sites satisfying the selection criteria is provided for each transcript. Clicking on this value generates an additional panel that provides the chromosomal location of each site along with the associated miRanda score and P-value. The target site panel also provides a list of all known miRNAs that target the selected transcript. The list of transcripts generated by miR-AT! can be sorted by either the cumulative score (default) or by using any of the other data columns, eg, gene name. The target list can be saved as a tab delimited text file. An additional feature of miR-AT! is the ability to generate histograms of target site score distributions. Histograms are provided for both individual scores and cumulative scores.
Functional analysis of identified targets
Microarray experiments often produce lengthy lists of differentially expressed genes. During the maturation of microarray technology it became apparent that such lists require computational tools that can identify biological processes and pathways that are associated with the genes in a given list. We have integrated miR-AT! with one of the most widely utilized and cited tools that belong to this class of bioinformatics resources: DAVID (Database for Annotation, Visualization and Integrated Discovery).22 DAVID allows for the rapid identification of enriched gene ontologies and biological pathways associated with a given gene list, and it also provides comprehensive functional annotation drawn from numerous biological databases. Among the resources available through DAVID are KEGG and Biocarta pathways. An output list of target genes in miR-AT! can be automatically submitted to DAVID by simply clicking the “Functional Annotation” button at the top of the output list. The automated miR-AT! procedure utilizes NCBI Entrez Gene IDs for submission to DAVID, and a new browser window will appear with DAVID results. Users are encouraged to explore the extensive help available at the DAVID web site (http://david.abcc.ncifcrf.gov/). Functional annotation clustering performed in this work was accomplished using the default settings of DAVID version 6.7. Ontology analysis utilized the GO_BP_ FAT records within DAVID, and the conservative EASE P-values were used.
Hierarchical clustering
To facilitate the identification of transcripts with similar microRNA target site patterns, we developed a novel adaptation of hierarchical clustering within miR-AT!. We integrated Java Treeview and Cluster 3.0 software libraries within the miR-AT! application.23,24 After using miR-AT! to identify predicted transcripts for a set of input microRNAs the user can press the “Clustering” button at the top of the page to produce a hierarchical clustering result. Predicted transcripts with similar patterns of target sites, considering the input microR-NAs, are clustered together. Quantitative values used in the clustering are derived from the scores of each microRNA/transcript combination. The clustering distance between two transcripts is determined by the correlation of microRNA target site scores in their 3′ UTRs. Additionally, microRNAs are clustered based on their target site patterns among all of the predicted targets, thus allowing for two-dimensional clustering. The clustering implementation in miR-AT! allows for the selection of a variety of distance metrics. The Spearman correlation distance metric was used in this work.
Analysis of signaling networks
To identify key nodes in signaling networks downstream of genes targeted by the collective group of microRNAs we utilized the ExPlain 3.0 analysis system (http://biobase-international.com/).25 This application requires a license and therefore is not integrated within miR-AT!. The key node analysis algorithm in the ExPlain system identifies downstream molecules that are connected to a maximal number of input molecules, within a specified distance. The default parameter settings and false discovery rate (FDR) computation were used.
Results
We used miR-AT! to predict the aggregate activity of six microRNAs that were repressed in metastatic breast cancer cells (hsa-mir-335, 126, 206, 122, 199a-3p, and 489) in the Tavazoie et al study. Focusing on target genes with multiple miRNA binding sites, we used the selectable parameters available in miR-AT! to identify transcripts having at least two miRNA binding sites representing at least two unique microRNAs from the input set. To avoid sites with low prediction confidence, we required each miRNA binding site to have a minimum MicroCosm score of 15. Analysis of nearly 900,000 target scores in MicroCosm revealed a near-normal distribution with a mean score of 16.398 and a standard deviation of 0.883 (data not shown). Based on the score distribution, a cutoff score of 15 eliminates the lowest 5% of scores from the MicroCosm database. Using miR-AT! with these parameter settings we identified 529 genes having multiple target sites of the six miRNAs. The list of transcripts is provided in Supplementary Table 1, ranked by cumulative score. One target transcript had six miRNA sites, four transcripts had five sites, 18 had four sites, 89 had three sites, and 417 had two sites. The cumulative scores for each transcript ranged from 30.4 to 95.1.
Predicted targets that have previously been confirmed
Among the list of combinatorial targets predicted by miR-AT! are genes associated with proliferation and metastasis and for which post-transcriptional regulation by at least one of the predicted microRNA interactions has previously been confirmed. Our combinatorial target analysis identified Aldolase A (ALDOA) as a putative target of miR-122 and miR-489. ALDOA is one of three aldolase isozymes which play a role in glucose metabolism. The dependency of proliferating tumors on elevated levels of glucose metabolism has been known for many years, and increased levels of aldolase in malignancies was first identified in 1953.26,27 Since then, numerous publications have reported elevated levels of ALDOA in a variety of cancers, including breast, hepatocellular, and lung.28–32 Increased levels of ALDOA have also been associated with invasiveness and metastasis.33,34 Consistent with the prediction obtained through miR-AT!, several labs have verified that ALDOA is a target of mir-122, and miR-122 expression results in a reduction of ALDOA mRNA levels.35–37 Our analysis indicates that miR-122 and miR-489 may cooperatively provide post-transcriptional regulation of Aldolase A and that their combined loss may contribute to dysregulation of Aldolase A expression.
General transcription factor 2B (GTF2B) is the third highest scoring target transcript of the collective group of six microRNAs. It is predicted by miR-AT! to be a target of microRNAs miR-122 (two sites), miR-126, miR-199a-3p, and miR-206, for a total of five sites. Validation of GTF2B regulation at the mRNA level by miR-122 has been reported in rat hepatocytes.36 Since GTF2B is a ubiquitous transcription factor involved in the regulation of many genes, concurrent repression of these four microRNAs could have extensive secondary effects on gene regulation.38
Functional annotation of the cumulative target gene set
We sought to identify genes associated with metastasis and proliferation from among the transcripts targeted by the collective set of the six miRNAs. An important feature of miR-AT! is the ability to seamlessly submit target gene lists to DAVID for functional annotation and analysis of enriched biological processes and pathways. The set of 529 genes identified with multiple target sites of the six miRNAs was submitted to DAVID for functional annotation using the automated hyperlink available in miR-AT!. Default DAVID settings were used. Of the input set, 414 genes were recognized in the DAVID database. Within DAVID we selected gene ontology biological process (GOTERM_BP_ FAT) to identify cellular processes associated with the genes targeted by the six microRNAs.
Several biological processes pertinent to proliferation and metastasis were flagged as statistically significant in the DAVID analysis, based on the number of target genes belonging to each category and compared to the number of genes in the given category found throughout the genome. Among the significant ontologies were cell cycle checkpoint (P = 0.0054) and negative regulation of cell adhesion (P = 0.018). Table 1 lists target genes associated with these categories and the microRNAs that target each gene.
Table 1.
Enriched biological processes among the 529 target genes having multiple microRNA binding sites.
| Entrez ID | Gene symbol | Gene name |
Number of microRNA sites |
|||||
|---|---|---|---|---|---|---|---|---|
| miR-335 | miR-126 | miR-206 | miR-122 | miR-199a-3p | miR-489 | |||
| Cell cycle checkpoint (P = 0.0054, 3.8-fold enrichment) | ||||||||
| 51499 | TRIAP1 | TP53 regulated inhibitor of apoptosis 1 | 1 | 1 | ||||
| 7272 | TTK | TTK protein kinase | 1 | 1 | ||||
| 1029 | CDKN2A | Cyclin-dependent kinase inhibitor 2A | 1 | 1 | ||||
| 60561 | RINT1 | RAD50 interactor 1 | 1 | 1 | ||||
| 8525 | DGKZ | Diacylglycerol kinase, zeta 104kDA | 1 | 1 | ||||
| 56984 | PSMG2 | Proteasome assembly chaperone 2 | 1 | 1 | ||||
| 11200 | CHEK2 | CHK2 checkpoint homolog | 1 | 1 | ||||
| 7040 | TGFB1 | Transforming growth factor, beta 1 | 1 | 1 | ||||
| Negative regulation of cell adhesion (P = 0.018, 5-fold enrichment) | ||||||||
| 7045 | TGFBI | Transforming growth factor, beta-induced, 68kDA | 1 | 1 | ||||
| 1029 | CDKN2A | Cyclin-dependent kinase inhibitor 2A | 1 | 1 | ||||
| 5921 | RASA1 | RAS P21 protein activator (GTPASE activating protein) 1 | 1 | 1 | ||||
| 395 | ARHGAP6 | RHO GTPASE activiating protein 6 | 1 | 2 | ||||
| 7040 | TGFB1 | transforming growth factor, beta 1 | 1 | 1 | ||||
Among the 529 predicted targets, five genes are involved in negative regulation of cell adhesion. Compared to the number of genes in the human genome that belong to this ontology, the frequency of occurrence in the target gene set represents 5-fold enrichment, as calculated in the DAVID functional annotation analysis. Several of these genes have been implicated in metastasis. TGFBI (transforming growth factor, beta-induced, 68kDA) is a gene that codes for an extracellular matrix (ECM) protein involved in cell adhesion and migration. Our results indicate that the 3′ UTR of TGFBI contains predicted binding sites for miRs 122 and 489, and that a decrease in expression of these two microRNAs may result in increased expression of TGFBI protein. Elevated expression of TGFBI has been associated with the progression and metastatic spread of human pancreatic cancer and hepatoma cells.39,40 Overexpression of TGFBI was demonstrated to significantly increase metastatic potential by promoting extravasation in colon cancer cells.41 TGFBI has also been associated with metastasis of esophageal squamous cell carcinoma.42
The biological process with the greatest statistical significance was cell cycle checkpoint (P = 0.0054). This category included eight genes from the set of 529 predicted targets and was enriched 3.8-fold, compared to the overall genome. A predicted target gene associated with metastasis and involved in both cell cycle checkpoint and negative regulation of cell adhesion is TGFB1 (transforming growth factor, beta1). This gene has two high-scoring target sites for miRs-122 and 199a-3p. While TGB1 inhibits early stage tumorigenesis, it promotes invasion and metastasis in later stages of the disease.43,44 Elevated plasma levels of TGFB1 are correlated with decreased survival in metastatic breast cancer patients.45 In a mouse xenograft model using metastatic MDA-MB-435 cells, knockdown of TGFB1 using siRNA resulted in a 90% decrease in the number of macroscopic lung metastases.46 The potential role of these two microRNAs in TGFB1 regulation warrants further investigation.
Interestingly, three transcripts associated with cell cycle checkpoint each have target sites for both miRs 335 and 489: TTK, TRIAP1, and RINT1. TTK is a dual-specificity kinase involved in centrosome duplication. Expression of TTK has been associated with cell proliferation.47 Excessive accumulation of TTK protein was linked to the production of extra centrosomes during mitosis and may lead to genomic instability and tumorigenesis.48 Dramatically increased TTK mRNA levels were found in genetically unstable breast cancer cell lines and in high-grade primary breast cancer tissue.49 Over-expression of TRIAP1 (Tp53 regulated inhibitor of apoptosis) has been reported to inhibit apoptosis induced by DNA damage,50 and elevated levels of TRIAP1 were found in greater than 50% of multiple myeloma cases.51 Our results indicate that concurrent loss of miRs 335 and 489 may result in dysregulation of this group of genes involved in cell cycle regulation.
Evaluation of statistical significance
We have relied on the established statistical model in DAVID to identify biological processes that are enriched with genes from the target gene set. The P-values provided by DAVID are derived using a modified Fisher exact test. The test determines if the proportion of genes from the target set belonging to a given gene ontology is significantly different than the proportion of genes from the overall genome belonging to the same ontology. The DAVID implementation of the Fisher Exact test (EASE score) subtracts one from the number of genes in an ontology category prior to calculating the significance, and is therefore more conservative than the standard Fisher test. To further assess the robustness of DAVID scores applied to microRNA target sets we sought to determine if the biological processes identified in our analysis of the six Tavazoie microRNAs would arise by chance when using randomly selected groups of microRNAs. We performed a bootstrap analysis with 100 trial sets, each comprised of six microRNAs randomly drawn from the MicroCosm database. For each trial the predicted target transcripts were identified with miR-AT! using the same parameter settings specified above, and the number of target genes associated with each gene ontology or biological process was tabulated. We then calculated an empirical P-value that represents the probability that the number of genes associated with a given ontology from the Tavazoie target set would occur by chance. The bootstrap P-value was obtained using pb = r/t where t is the total number of trials, and r is the number of trials that produced a gene count equivalent or greater to that obtained with the Tavazoie micro-RNAs for the given biological process.
We had previously noted that cell cycle checkpoint and negative regulation of cell adhesion are among the statistically significant ontologies identified by DAVID analysis of the 529 predicted targets of the six Tavazoie microRNAs (detailed above). From this target set, eight genes belong to the cell cycle checkpoint category, with a corresponding DAVID P-value of 0.0054. Of the 100 bootstrap trials the greatest number of target genes found in this category was five, providing a bootstrap P-value of zero. For the negative regulation of cell adhesion category five genes were originally identified among the 529 predicted targets, with a DAVID P-value of 0.018. Again, none of the bootstrap trials produced an equivalent or greater number of target genes for this ontology. Our bootstrap analysis clearly demonstrates that the number of genes from the Tavazoie set belonging to these ontologies is statistically significant and highly unlikely to have occurred by chance.
Using the statistical results obtained for all gene ontologies where more than one gene was identified from the Tavazoie target set, Figure 1 provides a comparison of the P-values from DAVID enrichment analysis to those derived from our bootstrap analysis. Each data point represents one gene ontology, and a clear trend is evident. For 96% of ontologies the DAVID P-value is greater than that obtained empirically with our bootstrap analysis, revealing that the DAVID values are indeed conservative and often underestimate significance of enrichment for microRNA target sets. We recognize that the bootstrap approach may offer greater statistical power, and implementation of this method within miR-AT! would be of value. However, the computational requirements necessary to accomplish bootstrap analysis for any possible combination of parameter settings and input microRNA set size precludes implementation at this time. We hope to offer this feature in a subsequent release of the software, but our results demonstrate that the enrichment P-values available through DAVID are reliable and generally underestimate significance.
Figure 1.
Comparison of enrichment P-values obtained from DAVID with those derived using bootstrap analysis for 900 gene ontologies. Bootstrap P-values were calculated from 100 trials using randomly selected sets of six microRNAs each. For each trial, target genes were predicted using miR-AT!, and the number of genes in each ontology category was tabulated. The bootstrap P-value represents the probability that random trials would produce an equivalent or greater number of genes for a given ontology than that obtained from the Tavazoie target set. DAVID P-values were obtained using default functional annotation settings in DAVID. The DAVID P-values consistently underestimate statistical significance of ontology enrichment for the microRNA target set.
Cyclin D1 and c-Myc are key nodes downstream of the cumulative set of genes targeted by the repressed microRNAs
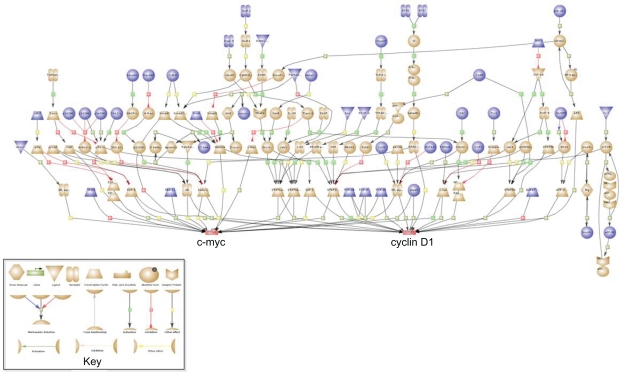
To detect central signaling nodes that may be focal points of dysregulation due to the collective loss of the six microRNAs, we identified converging signal transduction pathways downstream of the 529 predicted target genes. We used the key node search algorithm of the ExPlain 3.0 system (Biobase GmbH) to perform the analysis.25 This application requires a license and therefore is not integrated within miR-AT!. The ExPlain method explores cell signaling networks within a specified range from each input molecule (gene/protein) to find the most proximal molecule having the maximal number of connections to the overall set of input molecules. Since the resulting score may be influenced by the level of connectivity of each molecule, the total number of connections for each node is taken into account by the algorithm. Using the 529 predicted target genes as the input set and searching for key downstream nodes, the two highest scoring nodes were c-Myc and cyclin D1 (CCND1). c-Myc is downstream of 37 target transcripts and cyclin D1 is downstream of 36 target transcripts. The probability that these key nodes would achieve their scores by chance (false discovery rate) is 0.04 for c-Myc and 0.076 for cyclin D1. Figure 2 shows the network of target genes upstream of c-Myc and cyclin D1. Blue color indicates genes that are predicted targets of the six microRNAs. Supplementary Table 2 provides a list of the associated target genes.
Figure 2.
Cyclin D1 and c-Myc are key nodes downstream of the collective set of genes targeted by the six microRNAs. Analysis of convergent signaling pathways downstream of the 529 predicted target genes was performed using the key node algorithm of the ExPlain Analysis System (Biobase). Blue indicates transcripts that are predicted targets of two or more of the six repressed microRNAs. 37 targeted transcripts converge upon c-Myc and 36 converge upon cyclin D1, with some transcripts common to the two paths. Target genes are listed in Supplementary Table 2.
Amplification and over-expression of cyclin D1 and c-Myc are widely known to contribute to oncogenesis in a variety of tumors, including breast cancer, and is associated with decreased patient survival.52–57 A study using a mouse model of breast tumorigenesis demonstrated a synergistic interaction in response to concurrent over-expression of cyclin D1 and c-Myc, resulting in cells that were highly invasive and metastatic.58 Our computational analysis indicates that numerous genes targeted by the six repressed microRNAs are involved in signaling networks that converge upon cyclin D1 and c-Myc, potentially contributing in a focal manner to their dysregulation.
Clustering of transcripts by microRNAs target site patterns
As noted above, several target genes associated with cell cycle checkpoint each have target sites for both miRs 335 and 489. This observation raised the question as to whether some biological processes are under greater influence of specific combinations of microRNAs. To further explore combinatorial patterns of microRNA regulation in the cumulative set of target genes we implemented a novel adaptation of a clustering method frequently used in microarray gene expression analysis. Hierarchical clustering is widely used to identify subsets of genes having similar expression profiles when measured over a series of biological samples.59 This method is also commonly used to cluster biological samples that are similar based on gene activity assayed in a microarray experiment. Here, we utilized hierarchical clustering to identify clusters of target genes based on similarity of predicted microRNA target site patterns in their 3′ UTRs. For each of the 529 target transcripts, the total MicroCosm score of predicted target sites was tabulated individually for each of the six microRNAs. Transcripts with similar target site patterns were clustered together. Transcripts are represented as rows in our clustering results. This approach enabled us to identify clusters of genes that are similarly regulated by the six Tavazoie microRNAs, based on computational prediction of target sites. Additionally, we clustered the six microRNAs based on the target site tabulation, grouping together microRNAs that tend to co-occur within the same transcripts, as indicated by the columns in the clustering result.
The clustering result is shown in Figure 3, where rows represent the 529 target transcripts and columns represent the six microRNAs. The block representing each row-column intersection is colored to indicate the total score of target sites predicted within the transcript 3′ UTR for the given microRNA. Blue indicates no target site present, and red indicates a predicted target site. The red color intensity represents the cumulative score for all sites of the specified target-microRNA combination, with more intense color indicating a higher individual site score or multiple sites for the given microRNA. The dendrogram above the columns reveals that miR-122 and miR-206 are clustered together and is the most similar pair of microRNAs due to the coexistence of their sites in large number of target genes. The hierarchical clustering result shows that this group of transcripts is by far the largest sub-cluster associated with a pairwise combination of microRNAs.
Figure 3.
Hierarchical clustering of transcripts based on microRNA target site patterns. Red intensity reflects the total score of predicted sites for each microRNA/transcript combination. Brighter intensity indicates multiple target sites for a microRNA within a given transcript or a high scoring individual site. Blue indicates no predicted target site. Transcripts targeted by the miR-122/206 pair are enriched for genes associated with cytoskeleton organization and cell-matrix adhesion. Transcripts targeted by the miR-489/335 pair are enriched for genes associated with cell cycle control.
We then used miR-AT! to tabulate target genes having both sites for each pairwise microRNA combination. The results are summarized in Figure 4 and reveals that there are 174 transcripts having both miR-122 and miR-206 sites. This microRNA target site pair is found in nearly three times as many transcripts as the next most frequent pair of microRNAs. Inspection of the mature sequences for miRs 122 and 206 reveals that their seed regions are nearly identical; therefore, target sites for these two microRNAs tend to co-occur in the 3′ UTR. An alignment of microRNAs 122 and 206 was performed using the R-coffee algorithm which is suited for aligning non-coding RNA sequences (http://www.tcoffee.org).60 The alignment result shown in Figure 5 highlights the similarity between these two microRNAs. The seed region at the 5′ end of a microRNA is critical to its activity, and hybridization to a target transcript is greatly dependent on near-perfect base-pairing within this region.61 The eight base seed region of miRs 206 and 122 are identical with the exception of a single base difference which introduces an allowable G:U wobble. The G:U base pair occurs widely in RNA secondary structure.62 A number of confirmed miRNA target sites have been identified with G:U base pairs in the seed region, and it has been demonstrated that a G:U base pair can be tolerated within a 7mer or 8mer seed, albeit with some reduction in regulatory efficiency.6,61 The similarity of miRs 122 and 206 actually extends through the first ten nucleotides at the 5′ end, with a second G:U wobble found in the ninth position. These microRNAs are thus predicted to target an overlapping set of transcripts which would be dependent on the additive abundance of the two miRs. Based on our analysis, we propose that these two microRNAs could potentially act in a concerted and failsafe manner for many target genes, where the partial loss of one of the microRNAs might be compensated by the other microRNA in the pair. A loss of both microRNAs, as observed in the Tavazoie work, would result in extensive dysregulation of numerous target genes.
Figure 4.
Tabulation of the number of transcripts targeted by each pairwise combination of microRNAs. Transcripts were included in the tabulation if they have predicted sites for both microRNAs in the pair. The miR-122/206 pair is predicted to target 174 transcripts, far more than any other pair, and reflects the similarity in the seed regions for these two microRNAs. The targeted transcripts are predicted to be regulated by both microRNAs.
Figure 5.
Alignment of miRs 122 and 206 reveals extensive similarity in their seed regions, thereby accounting for the large number of transcripts co-targeted by this pair of microRNAs. Red indicates a perfect match, and yellow indicates an allowable G:U wobble. The ten bases in the 5′ seed region are highly conserved.
We applied miR-AT! and the integrated DAVID analysis to examine enrichment of biological processes associated with sets of genes targeted by pairwise combinations of the six microRNAs, which are reflected as subclusters in Figure 3. For the target gene set of each pairwise microRNA combination we identified enriched Gene Ontology Consortium biological process categories using the default statistical cutoff in DAVID and further filtering to identify categories with a minimum 2-fold enrichment. We found that the 174 transcripts targeted by both miRs 206 and 122 are enriched 5-fold for genes associated with regulation of cytoskeleton organization (P = 0.016) and 6-fold for cell matrix adhesion (P = 0.026). The 50 transcripts targeted by both miR-335 and miR-489 are 15-fold enriched with genes involved in cell cycle checkpoint (P = 0.016). The enriched sub-clusters are indicated in Figure 3.
Remodeling of the actin cytoskeleton is essential to migration and invasion of tumor cells.63,64 Five genes co-targeted by miR-122 and miR-206 are involved in this biological process, including ARPC3 (actin related protein 2/3 complex, subunit 3, 21 kDa). The ARP2/3 complex is part of the “minimum motility machine” that facilitates cell migration by creating cell protrusions driven through actin polymerization.65,66 This multiprotein complex is necessary for the formation of “invadopodia” protrusions that enable metastasizing cancer cells to invade the extracellular matrix and migrate into blood vessels during intravasation.67–69 Genes critical to the minimum motility machine were found to be dramatically up-regulated in invasive breast cancer cells that were selected using an in vivo invasion assay.66 Among this group was the p21 subunit of the ARP2/3 complex (ARPC3) which was found to be more than two-fold up-regulated. Our analysis suggests that the motile nature of the highly metastatic breast cells isolated in the Tavazoie work may be facilitated by the simultaneous loss of miRs 122 and 206 which are predicted to coordinately repress a number of genes involved in regulation of the actin cytoskeleton, including ARPC3. The loss of just one of these microRNAs may not be sufficient to significantly increase the level of transcripts which have both sites if sustained levels of the alternative microRNA are present. However, coincident loss of both microRNAs could result in a critical loss of post-transcriptional repression.
Thymosin beta 4 (TMSB4X) is a regulator of actin polymerization and has been implicated in tumor metastasis and cell motility. The TMSB4X transcript contains target sites for both miRs 122 and 206. A dramatic increase of invasiveness and motility has been observed in response to overexpression of TMSB4X in SW480 colon cancer cells.70 The same report also noted increased expression of TMSB4X in human liver metastases as compared to matched primary colorectal adenocarcinoma samples. Elevated expression of TMSB4X was correlated with metastatic potential in malignant mouse fibrosarcoma cells.71 Additionally, TMSB4X expression levels were found to be elevated in metastatic human melanoma cells, and overexpression was associated with increased tumor growth and lung metastases in a mouse melanoma model.72,73
Figure 4 reveals that the combination of miR-126 with any one of the other five microRNAs results in very few predicted targets having sites for both miRs. This appears to be due to the overall scarcity of miR-126 targets found throughout the genome. In the MicroCosm Targets database there are 212 human transcripts having at least one site with minimum score of 15.0 for miR-126. This compares to 976 targets for miR-335, 938 for miR-206, 1012 for miR-489, and 1091 for miR-199a-3p. Despite the relatively small set of target genes for miR-126, the loss of this microRNA appears to play a role in the metastasis of breast cancer. Tavazoie et al reported that the majority of primary tumors from breast cancer patients who relapsed demonstrated a loss of expression for miR-126.18 Investigating potential combinatorial activity of miR-126, an important target emerged from our computational analysis: RANKL. This gene codes for a member of the tumor necrosis factor (TNF) cytokine family and is known to induce osteoclast activity.74 Increased levels of RANKL have been associated with bone metastasis in a variety of cancers, and RANKL is actively being pursued as a therapeutic target.75–79 A recent study of renal cell carcinoma suggests that RANKL may be involved in metastasis to sites other than bone by stimulating cancer cell migration.80 Another study demonstrated that RANKL induced breast and prostate cancer cell migration.81 There are conflicting reports regarding RANKL expression in MDA-MB-231 cells. Some reports indicate that MDA-MB-231 cells alone in culture do not express RANKL but can induce RANKL expression through cell-cell contact with osteoblastic or stromal cells.82,83 However, a recent report found basal expression and secretion of RANKL in MDA-MB-231 cells using ELISA assays.84 It is currently unknown if microRNAs influence RANKL expression in tumor cells. Here, we find that four of the six Tavazoie microRNAs (miRs-126, 199a-3p, 335, 489) are predicted to target RANKL. Loss of expression for this collective set of microRNAs may result in RANKL dysregulation and over-expression. Additional investigations of RANKL regulation by these microRNAs are warranted.
Predicted microRNA activity is consistent with previously reported gene expression changes for PDGFA and KRT81
Since one of the modes of microRNA activity can produce changes in expression levels of target transcripts it is of interest to examine available expression data for predicted target genes. In miR-AT! we included an option to integrate gene expression data alongside microRNA target transcript predictions. The option is available in the results page following submission of a set of microRNAs and allows for import of a text file containing gene identifiers in the first column and quantitative expression values in the second column. The gene identifiers can be either NCBI Entrez or Ensembl identifiers. The quantitative values may be a ratio, fold change, or log ratio representing a change in transcript abundance.
In an earlier microarray report from the same laboratory that performed the Tavazoie et al microRNA study, a set of differentially expressed genes was identified in derivative MDA-MB-231 cells that produce aggressive lung metastases.85 Supplementary Table 2 from that publication provides gene expression changes for a select subset of genes identified as differentially expressed and having a minimum 3-fold change when comparing lung metastatic LM2 derivative cells and parental MDA-MB-231 cells. Since the same derivative cells were also used in the Tavazoie microRNA study we investigated if any of the predicted targets of the six down-regulated microRNAs were over-expressed in the highly metastatic derivatives, indicating potential microRNA activity at the transcriptional level. We cross referenced the Affymetrix probe identifiers from the supplemental table to Entrez Gene IDs using DAVID and created a tabular file with the Entrez IDs and reported fold changes. This file was imported into miR-AT! on the results page using the “Expression” button. The subsequent results displayed in miR-AT! revealed predicted targets with a corresponding expression change.
Several predicted target genes were over-expressed in the lung metastatic derivative cells, including PDGFA (platelet derived growth factor alpha polypeptide) and KRT81 (keratin, hair, basic 1; KRTHB1). The genes had 5.02 and 6.34-fold increased mRNA expression respectively in the highly metastatic cells as compared to parental cells (Minn et al Supplementary Table 2).85 Since these transcripts are also predicted to be targets of the repressed Tavazoie microRNAs, which were identified using the same cell lines, the evidence collectively suggest the increase in transcript level may be due to the loss of the microRNAs. PDGFA has one predicted miR-206 target site and two miR-122 sites, and is classified in the cell cycle control and cytoskeleton organization ontologies. PDGFA is expressed more frequently in breast tumors than in non-tumor breast tissue and also much more frequently in primary tumors with lymph node metastasis than in tumors from patients without metastasis.86 Elevated plasma levels of PDGF are correlated with decreased survival times of breast cancer patients and a greater extent of metastatic involvement.87,88 Inhibition of PDGF receptor signaling reduced tumor growth of human breast cancer cells implanted into mouse bone.89
The KRT81 gene encodes for a hair keratin protein and has predicted sites for miRs 335 and 206. This gene is normally expressed in the hair follicle; however, a 5′-truncated form of this gene (hHb1-ΔN) was found to be expressed in metastatic and primary breast carcinomas and is notably absent in non-malignant cells.90 Over-expression of the truncated form was also reported in four Epstein-Barr virus infected epithelial carcinoma cell lines.91 Using several breast cancer cell lines, it was determined that the truncated form of the gene was transcribed through an alternative promoter located in the fourth intron of the gene.92 The authors demonstrated that the protein product of hHb1-ΔN participates in cytoskeleton structure, and they suggested that it may alter the adhesive properties of cancer cells. Collectively, the previous reports from the Massague lab indicate that KRT81 gene expression was significantly elevated in both lung and bone metastatic cell derivates that were also used in the microRNA study. On the Affymetrix microarrays employed in the associated gene expression studies, the probe set targeting this gene (213711_at) hybridizes within exon 9 and would therefore measure expression of the full length and truncated transcripts. It is intriguing to consider that the reported increase of KRT81 messenger RNA levels in highly metastatic MDA-MB-231 cells reflects expression of the truncated transcript associated with malignancies. Since the truncation occurs at the 5′ end, it is expected that predicted microRNA binding sites for miRs 335 and 206 in the 3′ UTR would remain intact in hHb1-δN. Consequently, loss of these microRNAs may contribute to the over-expression of the truncated transcript which may affect cell adhesion in breast malignancies.
Discussion
Microarray and next generation sequencing technologies have enabled high-throughput analysis of microRNA expression profiles associated with disease progression. Numerous studies have found multiple microRNAs concurrently dysregulated in a variety of diseases. In these investigations, microarray assays are often followed by computational analysis to identify putative target genes of aberrant microRNAs. To date, most computational work has focused on individual microRNA-target interactions. However, recent reports have demonstrated cooperative and synergistic activity among microRNAs. Therefore, it is imperative to consider combinatorial activity when predicting the effect of multiple dysregulated microRNAs.
In this report we introduced a web-based tool, miR-AT!, that enables computational analysis of combinatorial microRNA activity. The application provides the ability to perform fully integrated pathway and ontology analysis of predicted target genes. Additionally, miR-AT! provides novel features for combinatorial analysis and clustering of transcripts using target site patterns. We applied miR-AT! to predict target interactions of six microRNAs that have been implicated in breast cancer metastasis.18 While the loss of individual microRNAs among this set may contribute towards metastatic progression, the consistent loss of the six microRNAs as a group in highly metastatic MDA-MB-231 cell derivatives suggests that cooperative activity may be involved in the aggressive behavior of these cells. Our in silico analysis identified numerous predicted combinatorial target genes previously implicated in cancer metastasis. The loss of only one of multiple microRNAs targeting a gene may not incur complete loss of suppression of the target, but coincident loss of multiple microRNAs could result in more dramatic changes in target transcript and/or protein levels.
Our analysis revealed that miR-122 and miR-206 are predicted to cooperatively target a large set of genes due to considerable similarity in their seed regions. The set of co-targeted transcripts is enriched for genes involved in cytoskeleton regulation and cell adhesion. The two microRNAs appear to function in tandem and perhaps offer a level of regulatory redundancy. Coincident loss of both microRNAs is predicted to result in extensive target gene dysregulation affecting cytoskeleton regulation and cell adhesion.
The metastatic spread of malignant cells is a complex process involving a number of steps, including local invasion, intravasation, extravasation, and remote colonization. These steps require dynamic activity in a range of cellular processes, and several are crucial to metastatic progression. A systems biology analysis of multiple cancer datasets identified cell cycle regulation, cytoskeletal organization, cell motility, antigen presentation, and energy metabolism as key processes perturbed in all metastatic tumors regardless of the primary tissue type.93 Our analysis revealed that the genes collectively targeted by the six microRNAs are significantly enriched for transcripts involved in the first three of these processes and thus may cooperatively contribute to the metastatic progression of breast cancer through the deregulation of these pathways. We also used cell network analysis to identify central signaling nodes downstream of the collective set of predicted target genes. The highest scoring nodes were cyclin D1 and c-Myc, indicating that the coincident loss of the six microRNAs may have a focal effect on these two important regulatory nodes, both implicated in oncogenesis and metastasis. These findings are consistent with observations that microRNAs play a vital role in regulating key pathways involved in metastasis.94,95
Our computational approach is a hypothesis generating method that enables the exploration of combinatorial microRNA activity. We have identified genes and cellular processes implicated in cancer metastasis that are predicted to be collective targets of the six dysregulated microRNAs. While beyond the scope of this report, additional investigations to characterize and validate these interactions are warranted. It is important to note that the predictions are based on computational models and potential false positives must be considered. However, our focus on target genes having multiple, high-scoring microRNA binding sites is expected to diminish false positives. Previously reported laboratory validation of some microRNA-target interactions identified in our analysis also provides confidence in the approach and the underlying prediction method.
Cooperative microRNA activity is likely an integral component of most cell regulatory networks, and the analysis of combinatorial effects may be of benefit in the development of miR-based therapeutic strategies. Both anti-miR and miR replacement approaches show promise, with the latter being of particular interest in cancer treatment since a number of studies have reported extensive repression of microRNAs in tumors.96 Combinatorial targeting may provide greater efficacy in repressing hyperactivated pathways than that which could be obtained through the use of a single microRNA.
Supplementary Material
Acknowledgments
In memorium of Anupam Bhattacharjee, a wonderful colleague who provided technical assistance. We would also like to thank Brad Sherman for assistance with DAVID integration, and Drs. Anton Enright and Stijn van Dongen for help regarding the use of the MicroCosm Targets database. This work was supported in part by NIEHS P30 ES06639, NSF CNS0521454 (H.J.), NSF IIS0612203 (H.J.), and the Wayne State University President’s Research Enhancement Program.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;2007(367) doi: 10.1126/stke.3672007re1. re1. [DOI] [PubMed] [Google Scholar]
- 2.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Molecular Cell. 2008;29(1):1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. microRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Wu L, Fan J, Belasco JG. microRNAs direct rapid deadenylation of mRNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(11):4034–9. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraldez AJ, Mishima Y, Rihel J, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 6.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes and Development. 2004 Mar 1;18(5):504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 8.Enright A, John B, Gaul U, Tuschl T, Sander C, Marks D. microRNA targets in Drosophila. Genome Biology. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z. Insight into microRNA regulation by analyzing the characteristics of their targets in humans. BMC Genomics. 2009;10:594. doi: 10.1186/1471-2164-10-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forrest AR, Kanamori-Katayama M, Tomaru Y, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2009 Dec 3; doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 11.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77(1):12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 12.Xin M, Small EM, Sutherland LB, et al. microRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009 Sep 15;23(18):2166–78. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gusev Y. Computational methods for analysis of cellular functions and pathways collectively targeted by differentially expressed microRNA: microRNAs: Part B. Methods. 2008;44(1):61–72. doi: 10.1016/j.ymeth.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Wang X. miRDB: A microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14(6):1012–7. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucl Acids Res. 2008;36(suppl_1):D159–64. doi: 10.1093/nar/gkm829. 2008/1/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiromatzo AO, Oliveira TY, Pereira G, et al. miRNApath: a database of miRNAs, target genes and metabolic pathways. Genet Mol Res. 2007;6(4):859–65. [PubMed] [Google Scholar]
- 17.Li L, Xu J, Yang D, Tan X, Wang H. Computational approaches for microRNA studies: a review. Mamm Genome. 2009 Dec 15; doi: 10.1007/s00335-009-9241-2. [DOI] [PubMed] [Google Scholar]
- 18.Tavazoie SF, Alarcon C, Oskarsson T, et al. Endogenous human microRNAs that suppress breast cancer metastasis. 2008;451(7175):147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucl Acids Res. 2006;34(Suppl_1):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA Targets. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008 Jan;36(Database issue):D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis G, Sherman B, Hosack D, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4(5):P3. [PubMed] [Google Scholar]
- 23.De Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004 Jun 12;20(9):1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 24.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004 Nov 22;20(17):3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 25.Zubarev RA, Nielsen ML, Fung EM, et al. Identification of dominant signaling pathways from proteomics expression data. J Proteomics. 2008 Apr 30;71(1):89–96. doi: 10.1016/j.jprot.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 27.Schade AL. Enzymic studies on ascitic tumors and their host’s blood plasmas. Biochim Biophys Acta. 1953;12(1–2):163–71. doi: 10.1016/0006-3002(53)90135-8. [DOI] [PubMed] [Google Scholar]
- 28.Shadidi M, Sorensen D, Dybwad A, Furset G, Sioud M. Mucosal vaccination with phage-displayed tumour antigens identified through proteomics-based strategy inhibits the growth and metastasis of 4T1 breast adenocarcinoma. Int J Oncol. 2008;32(1):241–7. [PubMed] [Google Scholar]
- 29.Asaka M, Alpert E. Subunit-specific radioimmunoassay for aldolase A, B, and C subunits: clinical significance. Ann N Y Acad Sci. 1983;417:359–67. doi: 10.1111/j.1749-6632.1983.tb32878.x. [DOI] [PubMed] [Google Scholar]
- 30.Asaka M, Nagase K, Miyazaki T, Alpert E. Radioimmunoassay of aldolase A. Determination of normal serum levels and increased serum concentration in cancer patients. Cancer. 1983;51(10):1873–8. doi: 10.1002/1097-0142(19830515)51:10<1873::aid-cncr2820511020>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 31.Castaldo G, Calcagno G, Sibillo R, et al. Quantitative analysis of aldolase A mRNA in liver discriminates between hepatocellular carcinoma and cirrhosis. Clin Chem. 2000;46(7):901–6. [PubMed] [Google Scholar]
- 32.Gure AO, Altorki NK, Stockert E, Scanlan MJ, Old LJ, Chen YT. Human lung cancer antigens recognized by autologous antibodies: definition of a novel cDNA derived from the tumor suppressor gene locus on chromosome 3p21.3. Cancer Res. 1998;58(5):1034–41. [PubMed] [Google Scholar]
- 33.Hamaguchi T, Iizuka N, Tsunedomi R, et al. Glycolysis module activated by hypoxia-inducible factor 1alpha is related to the aggressive phenotype of hepatocellular carcinoma. Int J Oncol. 2008 Oct;33(4):725–31. [PubMed] [Google Scholar]
- 34.Tanaka H. Determination of aldolase A by radioimmunoassay. Experimental and clinical studies on the diagnosis of cancer of the digestive tract. Hokkaido Igaku Zasshi. 1983 Jul;58(4):363–75. [PubMed] [Google Scholar]
- 35.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006 Feb;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Fabani MM, Gait MJ. miR-122 targeting with LNA/2’-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14(2):336–46. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elmen J, Lindow M, Silahtaroglu A, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008 Mar;36(4):1153–62. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tu K, Yu H, Hua YJ, et al. Combinatorial network of primary and secondary microRNA-driven regulatory mechanisms. Nucleic Acids Res. 2009 Oct;37(18):5969–80. doi: 10.1093/nar/gkp638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yue H, Yang B, Zhang H, et al. Clinical significance of TGF- beta1 and beta-glucuronidase synchronous detection in human pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2002 May;1(2):309–11. [PubMed] [Google Scholar]
- 40.Tang J, Zhou HW, Jiang JL, et al. BetaIg-h3 is involved in the HAb18G/CD147-mediated metastasis process in human hepatoma cells. Exp Biol Med (Maywood) 2007 Mar;232(3):344–52. [PubMed] [Google Scholar]
- 41.Ma C, Rong Y, Radiloff DR, et al. Extracellular matrix protein betaig-h3/TGFBI promotes metastasis of colon cancer by enhancing cell extravasation. Genes Dev. 2008 Feb 1;22(3):308–21. doi: 10.1101/gad.1632008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong FH, Huang CY, Su LJ, et al. Combination of microarray profiling and protein-protein interaction databases delineates the minimal discriminators as a metastasis network for esophageal squamous cell carcinoma. Int J Oncol. 2009 Jan;34(1):117–28. [PubMed] [Google Scholar]
- 43.Joshi A, Cao D. TGF-beta signaling, tumor microenvironment and tumor progression: the butterfly effect. Front Biosci. 15:180–94. doi: 10.2741/3614. [DOI] [PubMed] [Google Scholar]
- 44.Glick AB. TGFbeta1, back to the future: revisiting its role as a transforming growth factor. Cancer Biol Ther. 2004 Mar;3(3):276–83. doi: 10.4161/cbt.3.3.849. [DOI] [PubMed] [Google Scholar]
- 45.Ivanovic V, Demajo M, Krtolica K, et al. Elevated plasma TGF-beta1 levels correlate with decreased survival of metastatic breast cancer patients. Clin Chim Acta. 2006 Sep;371(1–2):191–3. doi: 10.1016/j.cca.2006.02.027. [DOI] [PubMed] [Google Scholar]
- 46.Moore LD, Isayeva T, Siegal GP, Ponnazhagan S. Silencing of transforming growth factor-beta1 in situ by RNA interference for breast cancer: implications for proliferation and migration in vitro and metastasis in vivo. Clin Cancer Res. 2008 Aug 1;14(15):4961–70. doi: 10.1158/1078-0432.CCR-07-4604. [DOI] [PubMed] [Google Scholar]
- 47.Mills GB, Schmandt R, McGill M, et al. Expression of TTK, a novel human protein kinase, is associated with cell proliferation. J Biol Chem. 1992 Aug 5;267(22):16000–6. [PubMed] [Google Scholar]
- 48.Kasbek C, Yang CH, Fisk HA. Mps1 as a link between centrosomes and genomic instability. Environ Mol Mutagen. 2009 Oct;50(8):654–65. doi: 10.1002/em.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan B, Xu Y, Woo JH, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006 Jan 15;12(2):405–10. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 50.Park WR, Nakamura Y. p53CSV, a novel p53-inducible gene involved in the p53-dependent cell-survival pathway. Cancer Res. 2005 Feb 15;65(4):1197–206. doi: 10.1158/0008-5472.CAN-04-3339. [DOI] [PubMed] [Google Scholar]
- 51.Felix RS, Colleoni GW, Caballero OL, et al. SAGE analysis highlights the importance of p53csv, ddx5, mapkapk2 and ranbp2 to multiple myeloma tumorigenesis. Cancer Lett. 2009 Jun 8;278(1):41–8. doi: 10.1016/j.canlet.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 52.Musgrove EA, Hamilton JA, Lee CS, Sweeney KJ, Watts CK, Sutherland RL. Growth factor, steroid, and steroid antagonist regulation of cyclin gene expression associated with changes in T-47D human breast cancer cell cycle progression. Mol Cell Biol. 1993 Jun;13(6):3577–87. doi: 10.1128/mcb.13.6.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schraml P, Kononen J, Bubendorf L, et al. Tissue microarrays for gene amplification surveys in many different tumor types. Clin Cancer Res. 1999 Aug;5(8):1966–75. [PubMed] [Google Scholar]
- 54.Bieche I, Olivi M, Nogues C, Vidaud M, Lidereau R. Prognostic value of CCND1 gene status in sporadic breast tumours, as determined by real-time quantitative PCR assays. Br J Cancer. 2002 Feb 12;86(4):580–6. doi: 10.1038/sj.bjc.6600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loden M, Stighall M, Nielsen NH, et al. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene. 2002 Jul 11;21(30):4680–90. doi: 10.1038/sj.onc.1205578. [DOI] [PubMed] [Google Scholar]
- 56.Al-Kuraya K, Schraml P, Torhorst J, et al. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004 Dec 1;64(23):8534–40. doi: 10.1158/0008-5472.CAN-04-1945. [DOI] [PubMed] [Google Scholar]
- 57.Kendall SD, Linardic CM, Adam SJ, Counter CM. A network of genetic events sufficient to convert normal human cells to a tumorigenic state. Cancer Res. 2005 Nov 1;65(21):9824–28. doi: 10.1158/0008-5472.CAN-05-1543. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Thakur A, Sun Y, et al. Synergistic effect of cyclin D1 and c-Myc leads to more aggressive and invasive mammary tumors in severe combined immunodeficient mice. Cancer Res. 2007 Apr 15;67(8):3698–707. doi: 10.1158/0008-5472.CAN-06-4000. [DOI] [PubMed] [Google Scholar]
- 59.Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001 Jun;2(6):418–27. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 60.Moretti S, Wilm A, Higgins DG, Xenarios I, Notredame C. R-Coffee: a web server for accurately aligning noncoding RNA sequences. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W10–3. doi: 10.1093/nar/gkn278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005 Mar;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varani G, McClain WH. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000 Jul;1(1):18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Machesky LM. Lamellipodia and filopodia in metastasis and invasion: Nuclear Dynamics and Cytoskeleton Signaling. FEBS Letters. 2008;582(14):2102–11. doi: 10.1016/j.febslet.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 64.Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE. Cell motility and cytoskeletal regulation in invasion and metastasis. J Mammary Gland Biol Neoplasia. 2007;12(2–3):143–52. doi: 10.1007/s10911-007-9046-4. [DOI] [PubMed] [Google Scholar]
- 65.Loisel TP, Boujemaa R, Pantaloni D, Carlier M-F. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401(6753):613–6. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 66.Wang W, Goswami S, Lapidus K, et al. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004 Dec 1;64(23):8585–94. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- 67.Weaver AM. Invadopodia: specialized cell structures for cancer invasion. Clin Exp Metastasis. 2006;23(2):97–105. doi: 10.1007/s10585-006-9014-1. [DOI] [PubMed] [Google Scholar]
- 68.Yamaguchi H, Lorenz M, Kempiak S, et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 2005 Jan 31;168(3):441–52. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J Cell Sci. 2009 Sep 1;122(17):3037–49. doi: 10.1242/jcs.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang WS, Chen PM, Hsiao HL, Wang HS, Liang WY, Su Y. Overexpression of the thymosin beta-4 gene is associated with increased invasion of SW480 colon carcinoma cells and the distant metastasis of human colorectal carcinoma. Oncogene. 2004;23(39):6666–71. doi: 10.1038/sj.onc.1207888. [DOI] [PubMed] [Google Scholar]
- 71.Kobayashi T, Okada F, Fujii N, et al. Thymosin-beta4 regulates motility and metastasis of malignant mouse fibrosarcoma cells. Am J Pathol. 2002;160(3):869–82. doi: 10.1016/s0002-9440(10)64910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nummela P, Yin M, Kielosto M, Leaner V, Birrer MJ, Holtta E. Thymosin beta4 is a determinant of the transformed phenotype and invasiveness of S-adenosylmethionine decarboxylase-transfected fibroblasts. Cancer Res. 2006 Jan 15;66(2):701–12. doi: 10.1158/0008-5472.CAN-05-2421. [DOI] [PubMed] [Google Scholar]
- 73.Cha HJ, Jeong MJ, Kleinman HK. Role of thymosin beta4 in tumor metastasis and angiogenesis. J Natl Cancer Inst. 2003;95(22):1674–80. doi: 10.1093/jnci/djg100. [DOI] [PubMed] [Google Scholar]
- 74.Fili S, Karalaki M, Schaller B. Mechanism of bone metastasis: the role of osteoprotegerin and of the host-tissue microenvironment-related survival factors. Cancer Lett. 2009 Sep 28;283(1):10–9. doi: 10.1016/j.canlet.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 75.Ohshiba T, Miyaura C, Inada M, Ito A. Role of RANKL-induced osteoclast formation and MMP-dependent matrix degradation in bone destruction by breast cancer metastasis. Br J Cancer. 2003 Apr 22;88(8):1318–26. doi: 10.1038/sj.bjc.6600858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michigami T, Ihara-Watanabe M, Yamazaki M, Ozono K. Receptor activator of nuclear factor kappaB ligand (RANKL) is a key molecule of osteoclast formation for bone metastasis in a newly developed model of human neuroblastoma. Cancer Res. 2001 Feb 15;61(4):1637–44. [PubMed] [Google Scholar]
- 77.Canon JR, Roudier M, Bryant R, et al. Inhibition of RANKL blocks skeletal tumor progression and improves survival in a mouse model of breast cancer bone metastasis. Clin Exp Metastasis. 2008;25(2):119–29. doi: 10.1007/s10585-007-9127-1. [DOI] [PubMed] [Google Scholar]
- 78.Hofbauer LC, Rachner T, Singh SK. Fatal attraction: why breast cancer cells home to bone. Breast Cancer Res. 2008;10(1):101. doi: 10.1186/bcr1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Santini D, Fratto ME, Vincenzi B, et al. Denosumab: the era of targeted therapies in bone metastatic diseases. Curr Cancer Drug Targets. 2009 Nov;9(7):834–42. doi: 10.2174/156800909789760375. [DOI] [PubMed] [Google Scholar]
- 80.Mikami S, Katsube K, Oya M, et al. Increased RANKL expression is related to tumour migration and metastasis of renal cell carcinomas. J Pathol. 2009 Aug;218(4):530–9. doi: 10.1002/path.2567. [DOI] [PubMed] [Google Scholar]
- 81.Jones DH, Nakashima T, Sanchez OH, et al. Regulation of cancer cell migration and bone metastasis by RANKL. Nature. 2006 Mar 30;440(7084):692–6. doi: 10.1038/nature04524. [DOI] [PubMed] [Google Scholar]
- 82.Park HR, Min SK, Cho HD, Kim DH, Shin HS, Park YE. Expression of osteoprotegerin and RANK ligand in breast cancer bone metastasis. J Korean Med Sci. 2003 Aug;18(4):541–6. doi: 10.3346/jkms.2003.18.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomas RJ, Guise TA, Yin JJ, et al. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology. 1999 Oct;140(10):4451–8. doi: 10.1210/endo.140.10.7037. [DOI] [PubMed] [Google Scholar]
- 84.Nicolin V, Narducci P. Soluble TRAIL could enhance bone destruction acting on Rank-ligand in estrogen-independent human breast cancer cell line MDA-MB-231. Acta Histochem. Mar;112(2):189–92. doi: 10.1016/j.acthis.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 85.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005 Jul 28;436(7050):518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anan K, Morisaki T, Katano M, et al. Vascular endothelial growth factor and platelet-derived growth factor are potential angiogenic and metastatic factors in human breast cancer. Surgery. 1996 Mar;119(3):333–9. doi: 10.1016/s0039-6060(96)80120-6. [DOI] [PubMed] [Google Scholar]
- 87.Ariad S, Seymour L, Bezwoda WR. Platelet-derived growth factor (PDGF) in plasma of breast cancer patients: correlation with stage and rate of progression. Breast Cancer Res Treat. 1991 Dec;20(1):11–7. doi: 10.1007/BF01833352. [DOI] [PubMed] [Google Scholar]
- 88.Seymour L, Dajee D, Bezwoda WR. Tissue platelet derived-growth factor (PDGF) predicts for shortened survival and treatment failure in advanced breast cancer. Breast Cancer Res Treat. 1993;26(3):247–52. doi: 10.1007/BF00665802. [DOI] [PubMed] [Google Scholar]
- 89.Lev DC, Kim SJ, Onn A, et al. Inhibition of platelet-derived growth factor receptor signaling restricts the growth of human breast cancer in the bone of nude mice. Clin Cancer Res. 2005 Jan 1;11(1):306–14. [PubMed] [Google Scholar]
- 90.Regnier CH, Boulay A, Asch PH, et al. Expression of a truncated form of hHb1 hair keratin in human breast carcinomas. Br J Cancer. 1998 Dec;78(12):1640–4. doi: 10.1038/bjc.1998.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nishikawa J, Kiss C, Imai S, et al. Upregulation of the truncated basic hair keratin 1(hHb1-DeltaN) in carcinoma cells by Epstein-Barr virus (EBV) Int J Cancer. 2003 Nov 20;107(4):597–602. doi: 10.1002/ijc.11289. [DOI] [PubMed] [Google Scholar]
- 92.Boulay A, Regnier CH, Anglard P, Stoll I, Tomasetto C, Rio MC. Transcription regulation and protein subcellular localization of the truncated basic hair keratin hHb1-DeltaN in human breast cancer cells. J Biol Chem. 2001 Jun 22;276(25):22954–64. doi: 10.1074/jbc.M101687200. [DOI] [PubMed] [Google Scholar]
- 93.Ptitsyn AA, Weil MM, Thamm DH. Systems biology approach to identification of biomarkers for metastatic progression in cancer. BMC Bioinformatics. 2008;9(Suppl 9):S8. doi: 10.1186/1471-2105-9-S9-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009 Oct 1;69(19):7495–8. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. Mar 15;126(6):1283–90. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trang P, Weidhaas JB, Slack FJ. microRNAs as potential cancer therapeutics. Oncogene. 2008 Dec;27(Suppl 2):S52–7. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.